Leishmaniasis is a group of endemic diseases in 98 countries, affecting approximately 12 million people, with approximately 350 million more at risk. These diseases are produced by a protozoan of the Leishmania genus with clinical manifestations in the skin, mucous membranes, and organs 1. The presentation of the disease, its evolution, and its response to treatment in the host vary according to the infective species. Therefore, identification of the diseases is relevant when determining the prognosis of the disease and when making an effective treatment choice. Species identification also allows for an understanding of the epidemiological behavior of the disease and contributes to the formulation of the control and prevention strategies appropriate to each region 2-5.
Currently, multilocus enzyme electrophoresis (MLEE) is considered the reference standard for the identification of Leishmania species due to its widespread use and ability to discriminate species in both the New and Old World 6-8. This methodology allows the characterization of the microorganisms by the electrophoretic mobility of a wide range of intracellular enzymes 9,10. Worldwide, nine reference centers provide species identification by MLEE, and only three of them are in the Americas 1. This, added to the complexity of the technique, limits the frequent application of this methodology to characterize Leishmania species in Colombia and the world 10-12.
Polymerase chain reaction (PCR) followed by an analysis of restriction fragment length polymorphism (RFLP) offers an alternative for identifying Leishmania species by analyzing the characteristic patterns of the different species obtained using restriction enzymes on amplified products of the genetic material of interest. This methodology has been expanded in the last two decades due to its speed in obtaining results, its ability to discriminate between species, and to adapt it to somewhat complex conditions regarding available technical resources, as well as the possibility of identifying Leishmania species directly from clinical samples 4,11,13,14.
For identifying Leishmania species via PCR- RFLP, different molecular targets have been used. Some of the most frequently used have been kinetoplast DNA (kDNA) 15-18, the GP63 gene 4, the β-tubulin gene 19, ITS rDNA 18,20, the miniexon gene 21-24, and the gene that encodes heat shock protein 70 (hsp70) 2,25).
Previous studies from our group have focused on identifying circulating species of Leishmania in Colombia by using MLEE, monoclonal antibodies and PCR with kDNA, ITS rDNA, miniexon and hsp70 as targets, with the last two being those that allow differentiation of all subgenus Leishmania and Viannia species circulating in Colombia 18,26-29.
Amplification of the miniexon gene permits to identify the species L. (L.) amazonensis, L. (L.) mexicana, and L. (L.) infantum by differences in amplified product size (22,23). When amplification is followed by restriction using the HaeIII enzyme, it is possible to identify L. (V.) braziliensis but not the complex L. (V.) panamensis/L. (V.) guyanensis22-24,30. The hsp70 gene amplification followed by RFLP with the BccI enzyme allows to differentiate between L. (V.) panamensis and L. (V.) guyanensis 25,31,32).
The aim of this study was to establish the concordance between PCR-RFLP using miniexon and hsp70 as molecular targets and the reference standard MLEE for the identification of circulating Leishmania species in Colombia.
Materials and methods
Type of design
We conducted a concordance study that used MLEE and PCR-RFLP for the identification of the Leishmania species circulating in Colombia.
Study population
Isolates came from patients diagnosed with cutaneous, diffuse and mucosal leishmaniasis who visited the Centro Dermatológico Federico Lleras Acosta E.S.E. between 1999 and 2011.
Sample
We included 96 samples out of the 800 available at the biological bank. The inclusion criteria was strains previously identified under the indirect fluorescent antibody test and classified like null phenotype.
Reference strains
The reference strains included L. (V.) braziliensis (MHOM/BR/75/M2903), L. (V.) panamensis (MHOM/PA/71/LS94), L. (V.) guyanensis (MHOM/BR/75/M4147), L. (L.) amazonensis (IFLA/BR/67/PH8), and L. (L.) mexicana (MNCY/BZ/62/M379). The electrophoretic and restricted patterns of the reference strains were used to determine the species of the isolates.
Parasite culture
Parasites were cultured in Senekjie’s medium and incubated at 26°C until we obtained a primary culture. Subsequently, a mass culture was performed using Schneider’s medium supplemented with 10% fetal bovine serum 33.
Species identification
Identification by MLEE and PCR-RFLP were performed from the same isolated sample. Samples were processed by different professionals for each of the tests. Isoenzyme identification was performed in a reference center of the Pan American Health Organization (PAHO).
Identification by MLEE
The preparation of the extract for analysis and the isoenzyme was performed according to the methods described by Saravia, et al. 34,35. Isoenzyme electrophoresis of the clinical isolates and reference strains was performed on a cellulose support according to the method described by Godfrey, et al. 36.
DNA extraction
The cetyltrimethylammonium bromide method was performed as described by Wagner, et al., in 1987 and used for both the reference strains and the clinical isolates 37.
Miniexon gene PCR
For gene amplification we used the primers reported by Marfurt, et al. (22,23). Each PCR reaction contained 20 mM Tris-HCl pH 8.4; 50 mMKCl; 1 mM MgCl2; 200 μMdATP, dCTP, dTTP and dGTP; 0.5 μM of each primer (Fme 5 ́-TAT TGG TATGCG AAA CTT CCG-3 ́ and Rme 5 ́-ACA GAA ACT GAT ACT TAT ATA GCG-3 ́) and DMSO 6%, 2 μl of DNA at 10 ng/μl and 0.05 units of Taq Polymerase Platinum (Invitrogen, São Paulo, Brasil) at a final volume of 50 μl.
Amplification was performed with the following conditions: An initial DNA denaturation at 95°C for 8 minutes; 40 amplification cycles of 95°C for 20 s, 55°C for 30 s and 72°C for 30 s; and a final extension cycle at 72°C for 5 minutes.
hsp70 gene PCR
Amplification was performed following the protocol proposed by García, et al. 25. Each PCR reaction contained 20 mMTris-HCl pH 8.4; 50 mMKCl; 2 mM MgCl2; 200 μMdATP, dCTP, dTTP and dGTP;0.4 μM of each primer (hsp70 sen 5’-GAC GGT GCC TGC CTA CTT CAA-3’ and hsp70 ant 5’-CCG CCC ATG CTC TGG TAC ATC-3’); 10% DMSO; 2 μl of DNA at 10 ng/μl and 0.05 units of Taq Polymerase Platinum (Invitrogen, São Paulo, Brazil) at a final volume of 50 μl.
Amplification was performed using the following protocol: An initial DNA denaturation at 94°C for 5 minutes; 33 amplification cycles at 94°C for 1 minute, 64°C for 1 minute and 72°C for 3 minutes; and a final extension cycle at 72°C for 10 minutes.
Visualization of the miniexon and hsp70 PCR products
Amplicons of the miniexon and hsp70 genes were analyzed by electrophoresis in 1% and 2% agarose gels, respectively. For the identification of species through PCR-RFLP, the molecular weights of the amplicons from the miniexon gene must be considered: 308 bp for L. (L.) mexicana, 283 bp for L. (L.) amazonensis, and 418 bp for the L. (L.) chagasi control, and between 255 and 227 bp for the Viannia subgenus (22, 23). The hsp70 gene presents a 1422-bp band for the Viannia subgenus 38.
Restriction fragment length polymorphism (RFLP)
Samples showing an amplification profile of the miniexon gene compatible with the Viannia sub-genus were subjected to restriction with the HaeIII enzyme (Invitrogen, São Paulo, Brazil).
The hsp70 gene of the samples that were identified as L. (V.) panamensis/L. (V.) guyanensis through the miniexon gene PCR-RFLP was also amplified and digested with the BccI enzyme (New England Biolabs, Ipswich, England).
Amplicons of the miniexon and hsp70 genes were purified using QIA quick PCR Purification kits (Qiagen, Düsseldorf, Germany). A total of 12.5 μl of purified PCR product was used for the digestion, with the addition of 1.4 μl of reaction buffer and 0.5 μl of restriction enzymes HaeIII (10 U/μl) and BccI (10 U/μl); the mix was incubated at 37°C for 120 minutes and at 45 volts.
Restriction products were analyzed by electrophoresis using high-resolution 2.5% agarose gels (Sigma-Aldrich, St. Louis, Missouri, USA) at 2.5 V/cm for 1.5 hours; a 100-bp molecular-weight marker was used (Bioline, London, England).
The samples that presented a restriction pattern of two bands of 108 and 118 bp with the HaeIII enzyme were identified as L. (V.) braziliensis. Amplicons that did not show a restriction pattern were identified as L. (V.) panamensis/L. (V.) guyanensis.
The restriction products obtained through the BccI enzyme in the reference strains L. (V) panamensis (MHOM/PA/71/LS94) and L. (V) guyanensis (MHOM/BR/75/M4147) showed the following patterns: Two bands of 428 and 890 bp and three bands of approximately 346, 428 and 544 bp. Species of clinical isolates were determined based on the restriction patterns obtained with reference strains.
Sequencing
We selected 80 isolates by simple random sampling and sequenced them to confirm the species identified by PCR and PCR-RFLP, including the samples identified as L. (V.) panamensis, L. (V.) braziliensis, L. (V.) guyanensis, and L. (V.) amazonensis.
For sequencing we used a BigDye Terminator, v3.1, Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an Applied Biosystems 3730xl DNA Analyzer system (Applied Biosystems, Foster City, CA, USA). For the analysis we used the software Geneious Pro 5.5.6 (Biomatters Ltda, Auckland, New Zealand) to align the sequences obtained with the sequences available at NCBI of the particular genes of the species of interest.
Statistical analysis
The results of the MLEE and PCR-RFLP of the species identified were recorded in an Excel data- base (Microsoft Office XP). Cohen’s kappa coefficient and the 95% CI were calculated using the software Stata®, version 13 (Stata Corp LP, College Station, Texas, USA) to determine the concordance between the MLEE and PCR-RFLP results.
Ethical considerations
This study was classified as safe and approved by Centro Dermatológico Federico Lleras Acosta E.S.E. Ethical Committee for the use of cryopreserved samples according to the ethical guidelines established by the Ethics Committee in Research, which are based on the Declaration of Helsinki and current Colombian regulations.
Results
The identification of the species for the isolates was performed according to the results obtained by PCR, PCR-RFLP and MLEE: L. (V.) braziliensis: 63.5%, L. (V.) panamensis: 21.9%, L. (V.) guyanensis: 5.2% and L. (L.) amazonensis: 9.4%. Sequencing confirmed the results obtained by PCR and PCR- RFLP. Figure 1 and figure 2A show PCR-miniexon gene and PCR-hsp70 gen amplification products from different Leishmania reference strains and clinical samples, respectively. The figure 2B shows restriction endonuclease DNA fragment patterns from amplicons of the PCR-hsp70 gene using BccI enzyme. Table 1 shows the identification results obtained by MLEE, miniexon gene PCR-RFLP, and hsp70 gene PCR-RFLP, and table 2 shows the results obtained by sequencing.

Figure 1 A. Line 1, molecular weight marker; Line 2, L. (V.) braziliensis reference strain DNA; Line 3, L. (V.) panamensis reference strain; Line 4, L. (V.) guyanensis reference strain; Lines 5-8, DNA from clinical samples of leishmaniasis cases typed as Leishmania Viannia subgenus; Lines 9-10 L. (L.) amazonensis reference strain; Lines 11-12, L. (L.) mexicana reference strain; Lines 13-14, L. (L.) infantum; Line 15, negative control

Figure 2 A. Line 1, molecular weight marker; Line 2, negative control; Line 3 L. (V.) panamensis reference strain DNA; Line 4, L. (V.) guyanensis reference strain DNA; Lines 5-8, DNA from clinical samples of leishmaniasis cases; Line 9, clinical sample from patient with suggested diagnosis of mucosal leishmaniasis and rhinoscleroma confirmed diagnosis. B. Line 1, molecular weight marker; Line 2, L. (V.) guyanensis reference strain; Line 3, clinical sample typed as L. (V.) guyanensis; Line 4, L. (V.) panamensis reference strain; Lines 5-6, clinical samples typed as L. (V.) panamensis
Table 1 Typing results of 96 isolates of Leishmania spp. by MLEE and PCR-RFLP methods
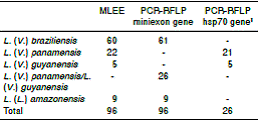
Ɨ:: Method applied only to 26 samples classified as L. (V.) panamensis/L. (V.) guyanensis by PCR-RFLP with the miniexon gene
Table 2 Typing results of 80 isolates of Leishmania spp. by MLEE and PCR-RFLP miniexon/hsp70 methods evaluated by sequencing

The concordance between the results of Leishmania species identification by PCR-RFLP and MLEE was “very good” according to the scale of Landis, et al. 39, for obtaining a kappa of 0.98 (CI95%: 0.98-1).
Among the 96 samples analyzed by both molecular tests (miniexon gene PCR-RFLP and hsp70 gene PCR-RFLP) and by MLEE, only one discordant result was observed. One of the isolates was identified through molecular tests as L. (V.) braziliensis but as L. (V.) panamensis by MLEE.
We obtained 96 isolates from patients whose ages ranged between 1.4 and 73 years and who had been diagnosed with cutaneous, diffuse cutaneous or mucosal leishmaniasis.
The patients came from 51 municipalities endemic for leishmaniasis in 15 different departments: Antioquia (n=1), Boyacá (n=4), Caldas (n=4), Caquetá (n=15), Casanare (n=2), Cundinamarca (n=24), Guainía (n=1), Guaviare (n=6), Meta (n=21), Nariño (n=1), Risaralda (n=1), Santander (n=4), Tolima (n=6), Valle del Cauca (n=2), and Vichada (n=4) (figure 3).
Discussion
According to the criteria of Landis, et al. 39, this study achieved a “very good” concordance between MLEE and PCR-RFLP of the miniexon and hsp70 genes for characterizing circulating Leishmania species in Colombia from clinical isolates. These results indicate that those obtained using PCR- RFLP for the species under study are equivalent to the results obtained using MLEE, which is considered the reference standard 1,6. The tests were applied on samples from the five geographical regions of the country, suggesting that it is possible to identify species despite the intraspecies variability associated with geographical distribution.
MLEE is a useful technique for characterizing species, as it permits the analysis of genetic variability between and among species, as well as their taxonomies and phylogenies 40,41. However, this methodology does not permit the direct identification of clinical specimens, the culture is required and it has low sensibility for samples from patients with mucosal and chronic cutaneous leishmaniasis, which prevents obtaining large amounts of parasites. Besides, MLEE is technically demanding and requires prolonged times for obtaining results 10-12,41. An alternative to discriminate between Leishmania species is PCR-RFLP, a widely used test that is performed with different molecular targets to determine the ability to differentiate species 42.
The miniexon and hsp70 genes were selected to identify circulating species in Colombia as they have several advantages: The number of copies of the genes allows for good test sensitivity, and these genes have been validated on a global scale with Old and New World species.
Recently, van der Auwera, et al., concluded after the analysis of several publications that the miniexon and hsp70 genes, compared to other molecular targets, exhibit the best resolution to identify medically relevant Leishmania species 43.
This claim is supported by experimental studies of the miniexon gene performed by Marfur, et al., as well as studies performed by Serin, et al., who evaluated the functionality of the miniexon gene as a phylogenetic marker of 17 species of the New and the Old World 22-24. The hsp70 gene has been evaluated for almost all circulating species in the world, with results that demonstrate its discriminatory capacity 28,32,44.
In our study, we identified L. (V.) braziliensis using PCR-RFLP, and L. (L.) amazonensis using only PCR based on amplicon size, results that were confirmed by sequencing. In Colombia, even though the prevalent species are associated with the Viannia subgenus, L. (L.) amazonensis recently was associated with cutaneous, diffuse cutaneous, and mucosal leishmaniasis. PCR of the miniexon gene allows the identification of L. (L.) amazonensis, L. (L.) mexicana, and L. (L.) chagasi only by amplicon size. This approach is very useful and less expensive than other methods that require additional tests such as RFLP or sequencing.
The hsp70 gene has also been extensively evaluated in Old and New World species. García, et al., were the first to identify its functionality to diagnose and differentiate species from the sub- genera Leishmania and Viannia, complemented with techniques such as RFLP and sequencing 29,32,38,45-48.
This technique has also proven to be useful in differentiating species from the L. guyanensis complex, which includes L. (V.) panamensis and L. (V.) guyanensis. These species are of great clinical and epidemiological interest in Colombia because L. (V.) guyanensis was associated with the largest epidemic of leishmaniasis in the country. This species also presents differences in the in vitro susceptibility response to drugs compared to other species of the Viannia subgenus 49,50.
In our study, hsp70 gene amplification and RFLP using the enzyme BccI were very useful, as they allowed discrimination between L. (V.) panamensis and L. (V.) guyanensis. The results obtained in the concordance analysis between the two tests suggest that they are equivalent for identifying the species circulating in Colombia. Our results are consistent with those obtained by da Silva, et al., who found an overall concordance between MLEE and PCR- RFLP of the hsp70 gene of the circulating species in Brazil, based on which the authors concluded that the latter method can replace MLEE in the routine identification of Leishmania species 51.
Another method recently used to identify species is the high-resolution analysis of dissociation using the hsp70 gene, which has demonstrated good agreement with PCR-RFLP and MLEE 52. Cardoso da Graça, et al., compared the molecular targets ITS, G6PD and hsp70 with MLEE, and found very good concordance between the methodologies for the identification of L. (L.) amazonensis, L. (L.) infantum, L. (V.) guyanesis, L. (L.) lanzoni, L. (V.) naiffi, and L. (L.) shawi, but discordance for the identification by MLEE of L. (V.) braziliensis53, which was the only dissenting finding of the current study.
Several hypotheses have been proposed to explain the difference between the identification results of the various methodologies, including the possibility of mixed infection, variation in the patterns according to the markers used, and the similarity between species of the Viannia subgenus 53,54.
The results indicate that the PCR-RFLP of the miniexon and hsp70 genes permit the identification of circulating species in the country. This test could be used in more laboratories compared to MLEE because it is technically simpler and the cost is 50% less than for MLEE. This work includes the largest number of samples reported in the country, identified by molecular tests and with a wide geographical distribution.
The test is restricted to native species, but it would be possible to expand its spectrum using other restriction enzymes to identify new circulating or imported species. Here we propose an identification algorithm for the Leishmania species that are circulating in Colombia (figure 4).
















