It is well known that the use of broad-spectrum antibiotics such as aminoglycosides, third- and fourth-generation cephalosporins, and fluoroquinolones allows selecting opportunistic multidrug-resistant bacteria which can colonize inpatients and, in some cases, cause serious infection difficult to treat 1,2. As in many warm-blooded animals, human intestine is known to harbor bacteria capable of causing infections in hospitals and in the community 3,4.
In the natural environment, the selection of multidrug-resistant bacteria may occur due to the production of antibiotic compounds by soil bacteria, but also due to discharges of human and animal feces that may contain antimicrobial drug residues 5,6. Some resistance encoding genes may be chromosomal housekeeping genes involved in the bacterium own protection against the antimicrobial compounds they produce by themselves or by other bacteria in the surrounding environment 6,7.
Additionally, some antimicrobial drugs are still used in animal farms and aquaculture as growth promoters and as prophylactic agents, which can also cause an artificial selection of antimicrobial-resistant bacteria 8-10. The use of antimicrobial drugs in humans, animal husbandry and veterinary care may enable the selection of bacterial strains with potential resistance traits resulting in host colonization and infection by different bacteria, such as those belonging to the Enterobacteriaceae family. This event may result in the appearance of bacteria with different resistance profiles which challenge treatment 11,12.
Another major concern is the existence of mobile genetic elements in some bacteria such as plasmids, gene cassettes, and transposons that can assemble blocks of resistant genes and transfer them horizontally enabling other bacteria to acquire genes encoding for resistance mechanisms to several antimicrobial drugs 13-15.
The ecosystem of several bodies of water has been seriously compromised by human practices and lifestyle. The discharges of households, hospitals and industrial residues without previous appropriate treatment have added to the problematic of bacterial antimicrobial resistance dissemination throughout these environments, which are used by humans and animals in many ways: for sports, professionally or for survival, users who may become colonized or infected by resistant bacteria 16-18.
Guanabara Bay is the second largest bay in Brazil (with 380 km2); it runs all along Rio de Janeiro urban cost line and has suffered intense degradation for many years due to population growth and industry development 19. Among the consequences of the degradation suffered by the accumulation of waste and sewage in these areas, we can mention the floods during heavy rain periods and the diseases caused by various microorganisms they brought with them.
In a previous study, Meirelles-Pereira, et al., detected the presence of strains of multidrug-resistant bacteria in sewage samples from a hospital in Rio de Janeiro, as well as from coastal lagoons in the city 20. The presence of extended-spectrum beta-lactamases (ESBL) and enzymes modifying aminoglycosides (EMA) was confirmed by phenotypic and PCR assays (data not shown).
The purpose of this work was to identify specific genes encoding EMA, ESBL, and plasmid mechanisms of quinolone resistance (PMQR) in Klebsiella pneumoniae subsp. pneumoniae, Escherichia coli, and K. pneumoniae subsp. ozaenae strains, which were isolated from Guanabara Bay water samples and hospital waste to better understand the dissemination of resistant mobile genetic elements in the natural environment and in hospital settings.
Materials and methods
Study area
The Guapimirim River, which is part of the Magé Channel, supplies the municipalities of São Gonçalo and Niterói and is located on the northern coast of the hydrographic region of the Guanabara Bay where the best preserved rivers are located 19.
The Guaxindiba River, located on the eastern coast, is born in São Gonçalo and it traverses part of the largest preserved mangrove area of the Guanabara Bay (the Guapimirim Environmental Protection Area) before discharging into the bay in its lower section. Large masses of sewage and other waste produced by the population inhabiting its surroundings are deposited in its waters through tributaries contributing to the degradation of the mangroves.
The Caceribu River is one of the main contributors to the Guanabara Bay. With almost 60 km of extension, its springs originate in the mountains of Rio Bonito and Tanguá municipalities and flow on the eastern slope of the Guanabara Bay through the Guapimirim mangrove 19.
The Mangue Channel on the western coast and its tributaries run through the most densely populated areas and they receive large quantities of treated and untreated sewage and residues including antimicrobials from industries and surrounding residential areas. The waters in the region comprising the Channel between Governor and Fundão Islands and the mainland have the worst quality, with high concentrations of nutrients such as phosphorus and nitrogen, as well as fecal coliforms, and a low concentration of oxygen due to the releases of raw or partially treated effluents from industrial and residential areas located north of Rio de Janeiro 19.
Strain selection
We studied 37 strains: 26 of E. coli, nine of K. pneumoniae subsp. pneumonia, and two of K. pneumoniae subsp. ozaenae. These strains were isolated from water collected in rivers that flow into the Guanabara Bay and from clinical specimens collected at a university hospital in Rio de Janeiro.
Environmental samples were collected at Guapimirim, Caceribu, and Guaxindiba Rivers and in the Magé Channel (Guapimirim Environmental Protected Area - Magé, RJ) in April, 2009 (figure 1). We also collected samples from two different areas of the Fundão Island rivers, from Bica beach (Governador Island), and the Mangue Channel in July, 2009. Regarding clinical samples, the criterion for selection was growth in selective media. Residues of hospital origin containing antimicrobials and microorganisms are continuously released into aquatic receptacles contaminating the environment and favoring the dissemination of transferable resistance markers between the microorganisms.
Isolation of strains from clinical material
Strains of K. pneumoniae and E. coli from different clinical materials collected and analyzed in the bacteriology laboratory of the university hospital between May and July, 2010, were inoculated in eosin methylene blue agar (EMB) (Oxoid) containing 8 μg/mL of gentamicin (Sigma) and those with positive growth were selected for the study.
Isolation of strains from water samples
Sterile flasks were used to collect the water samples, which were then transported on ice to the laboratory in the next six hours and tested straight after arrival according to the Environmental Protection Agency (EPA) protocols 21. A selection criteria was resistance to gentamicin as for the clinical samples, but also resistance to cephalotin, the latter determined by adding 32 μg/mL of cephalotin (Sigma) into the EMB media. The collection was done during low tide to determine the effect of higher hydrolytic pressure in the drained areas under study. The strains with macroscopic characteristics compatible with the bacterial species of interest were identified according to Winn, et al.22.
Antimicrobial susceptibility tests
The antimicrobial susceptibility tests (AST) was carried out using the agar diffusion method according to the Clinical Laboratories Standards Institute (CLSI) 23. Escherichia coli strain ATCC 25922 was used as control.
The following antimicrobial agents (Oxoid) were used with their respective potencies: Cotrimoxazol (Sut- 25 μg), cephalotin (Cph- 30 μg), ceftazidime (Caz- 30 μg), cefoxitin (Fox- 30 μg), cefotaxime (Ctx- 30 μg), cefuroxime (Crx- 30 μg), ceftriaxone (Cro- 30 μg), cephazoline (Cfz- 30 μg), aztreonam (Atm- 30 μg), ertapenem (Etp- 10 μg), imipenem (Ipm- 10 μg), cefepime (Cpm- 30 μg), piperacillin/tazobactam (Ppt- 100/10 μg), amoxillin/clavulanic acid (Amc- 20/10 μg), ampicillin/sulbactam (Asb- 10/10 μg), chloramphenicol (Chl- 30 μg), ciprofloxacin (Cip- 5 μg), norfloxacin (Nor- 10 μg), tetracycline (Tet- 30 μg), ampicillin (Amp- 10 μg), gentamicin (Gen- 10 μg), amikacin (Ak- 30 μg), kanamycin (Kan- 30 μg), tobramycin (Tob- 10 μg).
Phenotypic detection of ESBL
The isolates resistant to second and third cephalosporin’s generation in AST were submitted to confirmatory tests of ESBL production by the double-disc synergy test and the approximation test following the CLSI standards 24. Klebsiella pneumoniae strain ATCC 700603 was used as the test control.
Plasmid extraction
Plasmid DNA alkaline extraction was performed on all strains following Gonçalves, et al.25. The strains were subjected to electrophoresis in 0.8% agarose gel (Sigma) for plasmid detection. We used the E. coli R861 strain plasmid DNA as weight marker and the E. coli K12 R23 as the negative control.
Gene detection by PCR
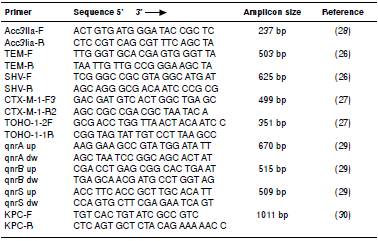
PCR was performed using primers compatible with ESBL, EMA, and PMQR in all K. pneumoniae subsp. pneumoniae, K. pneumoniae subsp. ozaenae, and E. coli strains resistant to any antimicrobial drug tested in the AST following the work developed by Arlet, et al.26, Pitout, et al.27, van de Klundert, et al.28, and Jiang, et al.29.
Isolates showing resistance to carbapenems were sent to the Healthcare Associated Infections Laboratory (LAPIH) of the Oswaldo Cruz Institute (Fiocruz) for the identification of the blaKPC gene according to Yigit, et al. 30.
The thermal cycling conditions were performed in a Cetus model 480 thermal cycler (Perkin-Elmer, Norwalk, CT).
PCR assays were performed in a 25 μl total volume adding the following components to the reaction tubes: 1 μl of target DNA (obtained from dilutions of colonies in 50 μl of 10 mM Tris, 1 mM EDTA - pH 8.0 (Sigma), 1.5 mM MgCl2 (Invitrogen), 0,2 mM of dNTP mixture (dATP, dTTP, dCTP and dGTP) (Invitrogen), 20 pmol of each primer (Invitrogen), 1x PCR buffer (Promega), and 1.25 U of Taq DNA Polymerase (Invitrogen).
The amplified products were subjected to electrophoresis in a 1.5% agarose gel (Sigma). Table 1 shows all primers used in the PCR reactions and the expected amplicon sizes.
Results
Strain isolation from clinical material
We isolated five strains of K. pneumoniae subsp. pneumoniae and five strains of E. coli from the EMB media containing gentamicin.
Strain isolation from water samples
We isolated 17 E. coli, four K. pneumoniae subsp. pneumonia, and one K. pneumoniae subsp. ozaenae from the media containing gentamicin and four E. coli and one K. pneumoniae subsp. ozaenae from the media containing cephalotin.
Antimicrobial susceptibility testing and ESBL confirmation test
To determine the resistance profiles of all isolated strains we used different classes of antimicrobial drugs ranging from those first used in therapeutics to the latest introduced in the medical practice.
There were some differences between strains isolated from media supplemented with gentamicin and those isolated from media supplemented with cephalotin in the water samples. Of the isolates obtained from gentamicin media, eight (36.4%) were resistant to cephalotin on the AST and of those obtained from cephalotin media, two (40%) were resistant to gentamicin on the AST. In water isolates, ten (37%) showed multidrug-resistance profiles similar to those displayed by clinical isolates, i.e., they were resistant to drugs used specifically in healthcare settings.
Twelve isolates were tested to confirm the presence of ESBL enzymes and 11 (91.7%) of them were positive. Fourteen (51.8%) showed resistance to ciprofloxacin and norfloxacin, four (14.8%) to amikacin, and one from Caceribu River and one from the Mage Channel were resistant to cefepime, a fourth generation cephalosporin usually used exclusively in hospitals. Furthermore, four isolates from Bica beach, one from the Mangue channel, and two from Fundão Island II were all resistant to cefepime. One K. pneumoniae subsp. ozaenae isolate, also from Fundão Island II, was resistant to ertapenem.
Table 2 presents information regarding water isolates, such as the collection site, the most relevant environmental parameters, and the antimicrobial resistance profile.
Table 2 Enterobacteriaceae isolated from water on the different collection sites at the Guanabara Bay including their resistance profiles
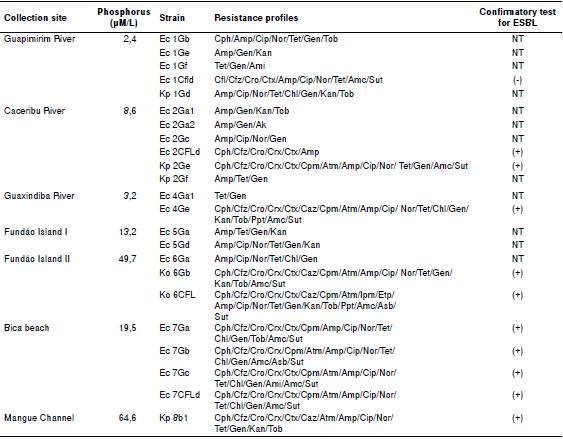
Ec: E. coli; Kp- K. pneumoniae; Ko: K. pneumoniae subsp. ozaenae; Cph: Cephalotin; Cfz: Cephazolin; Cro: Ceftriaxone; Crx: Cefuroxime; Ctx: Cefotaxime; Caz: Ceftazidime; Cpm: Cefepime; Atm: Aztreonam; Ipm: Imipenem; Etp: Ertapenem; Amp: Ampicillin; Cip: Ciprofloxacin; Nor: Norfloxacin; Tet: Tetracycline; Chl: Chloramphenicol; Gen: Gentamicin; Ak: Amikacin; Kan: Kanamycin; Tob: Tobramycin; Ppt: Piperacillin/tazobactam; Amc: Amoxillin/clavulanic acid; Asb: Ampicillin/sulbactam; Sut: Cotrimoxazol; NT: not tested
Regarding the clinical isolates, eight (80%) showed a multidrug-resistant profile typical in hospital settings. All ten isolates were resistant to amikacin, five (50%) were sensitive to chloramphenicol, six (60%) exhibited resistance to ciprofloxacin and norfloxacin, and four (40%) isolates were resistant to cefepime. Additionally, three (30%) isolates showed resistance to imipenem and ertapenem, which may indicate carbapenemases production, and five (50%) indicated ESBL production.
Table 3 shows information regarding clinical isolates, such as the sample type, the hospital ward, and the antimicrobial resistance profile.
Table 3 Enterobacteriaceae isolated from clinical samples including their resistance profiles
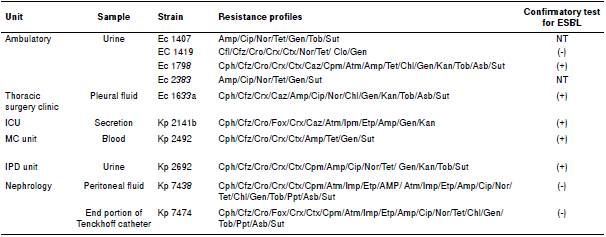
Ec: E. coli; Kp: K. pneumoniae; ICU: Intensive care unit; MC: Medical clinic unit; IPD: unit of infectious-parasitic diseases; Cph: Cephalotin; Cfz: Cephazolin; Cro: ceftriaxone; Fox: Cefoxitin; Crx: Cefuroxime; Ctx: Cefotaxime; Caz: Ceftazidime; com: Cefepime; Atm: Aztreonam; Ipm: Imipenem; Etp: Ertapenem; Amp: Ampicillin; Cip: Ciprofloxacin; Nor: Norfloxacin; Tet: Tetracycline; Chl: Chloramphenicol; Gen: Gentamicin; Ak: Amikacin; Kan: Kanamycin; Tob: Tobramycin; Ppt: Piperacicllin/tazobactam; Amc: Amoxillin/clavulanic acid; Asb: Ampicillin/sulbactam; Sut: Cotrimoxazol; NT: not tested
Plasmid DNA extraction
All strains isolated from water and nine (90%) strains isolated from clinical samples had at least one plasmid band.
Polymerase chain reaction (PCR)
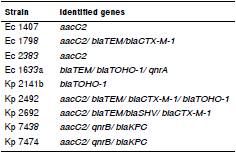
Of all the water isolates collected at the Guanabara Bay, nine (37.5%) showed PCR amplicon specific to EMA, two (18.2%) only for ESBL, and one (16.6%) for PMQRs. Seven (63.3%) isolates had positive amplification for both EMA and ESBL encoding genes. Two (7.4%) showed the presence of EMA, ESBL, and PMQR encoding genes (table 4).
Table 4 Genes found by PCR in the Enterobacteriaceae strains isolated from water samples
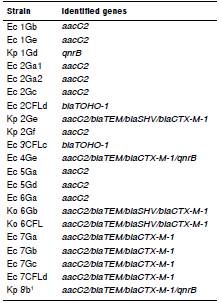
Ec: E. coli; Kp: K. pneumoniae; Ko: K. pneumoniae subsp. ozaenae
Of the clinical isolates, two (20%) showed gene amplification for EMA and only one (20%) for ESBL. Three (60%) isolates presented an amplicon for both EMA and ESBL; one (16%) for ESBL and PMQR, and two (20%) had PCR positive results for all EMA, ESBL, and PMQR encoding genes (table 5).
Discussion
The number of bacterial strains resistant to antimicrobial drugs has been increasing considerably in the natural environment due to the vast use of these drugs in hospitals and in the community, as well as in animal husbandry and veterinary care. The problem has increased due to hospital sewage dumping in natural environments, particularly in bodies of water, which facilitates the dissemination of bacterial strains harboring resistant and virulent mechanisms throughout them and may contribute to an outbreak of massive proportions, as well as the dissemination of mobile genetic elements carrying resistant genes and concomitant virulent genes.
Water sample collection was performed in two different areas of the Guanabara Bay which are substantially distinct from each other as regards their ecology and preservation practices and legislation. In the protected area of Guapimirim, the quality of water is supposed to be good for human consumption and recreation and the mangrove should be well preserved. Differently, the Fundão and Governador Islands are known to be strongly affected by massive discharges of treated and untreated human and animal sewage due to the vast population living in their surroundings.
No differences were noticed between the resistance profiles in the isolates from the Guapimirim area, Fundão and Governador Islands, and the Mangue Channel, although Guapimirim isolates showed less diverse resistance profiles in the different antimicrobial classes. In all the collection sites within the Guapimirim area, there was at least one isolate showing a resistance profile compatible with hospital strains which are known to be exposed to stronger antimicrobial drugs.
According to Negreiros, et al. 31, population growth and industry development, including pharmaceutical industries, affecting these bodies of water have contributed to their degradation. The authors also state that there is a massive agricultural activity in the vicinities of these bodies of water. Besides its privileged geographical location, these areas have been affected by the lack of sewage treatment infrastructures and policies unable to fulfill the demand of population growth. The majority of multidrug-resistant strains were, as expected, isolated in the areas with higher human impact.
It is not possible to discard that bacteria with resistant genes enclosed in plasmids may be disseminated to the bodies of water and that humans might be colonized with these strains when they use the water for consumption, recreational or professional reasons 32,33. Prado, et al. 34, demonstrated the presence of K. neumonia multidrug-resistant strains in water samples collected from hospital sewage treatment facilities. They found that almost 50% of the isolates were positive for beta-lactamic enzyme production and, within this group, 70% were resistant to gentamycin and 10% to ciprofloxacin. In the present study, 80% of the water isolates were positive for E. coli and of these, 52% was collected in the Guapimirim protected area and, besides, some multidrug-resistant isolates were found, which may indicate that these areas are being subjected to untreated dumping.
Regarding the clinical isolates, the use of selective agar media supplemented with gentamycin did not allow for the isolation of an enormous amount of isolates, although a high percentage of multidrug resistance was observed. According to Magiorakos, et al. 35, resistance to gentamycin may indicate a multidrug-resistant profile.
The water isolates from all the sites exhibited plasmids with different weights, even those strains with intrinsic resistance which were not submitted to antimicrobial pressure. Likewise, clinical isolates presented plasmids with different weights. Vaydia’s 14 study on E. coli and K. neumonia of clinical origin containing plasmids and showing resistance to beta-lactamic and other antimicrobials identified by conjugation the transference of genes encoding those traits.
In this study, we detected by PCR the presence of genes encoding resistance to the antimicrobial groups proposed for analysis. The water isolates Ec 4Ge and Kp 8Gb1, collected at Mage and Mangue Channels showed genes encoding for resistance to the three groups of antimicrobial drugs, as well as the isolates Kp 7438 and Kp7474 collected from peritoneal fluid and the exit site of Tenckhoff catheter, respectively.
Previous studies have described the existence of Enterobacteriaceae strains isolated from food, water, and clinical specimens associated to human infections, which harbor genes encoding resistance to different antimicrobial classes, as well as genes encoding for the same antimicrobial class. Ryu, et al. 36, reported on the presence of integrons containing genes which encode resistance to ESBLs, EMAs, and tetracycline in E. coli strains isolated from meat fish and seafood in Korea. In clinical specimens in Korea, Shin, et al. 2, isolated E. coli and K. neumonia strains resistant to aminoglycosides (including gentamycin) harboring genes encoding for ESBLs and PMQRs associated with integrons enclosed in plasmids transferable by conjugation.
Gene aacC2 was detected in 58.3% of the water isolates and 70% of the clinical isolates. Gonçalves, et al. 37, detected this gene in clinical isolates containing a transposon and insertion sequences (IS), which may indicate that this gene is vastly disseminated in hospital settings, water, soil, and natural environment. Genes found in bacteria isolated from water and animals are similar to those found in pathogenic bacteria isolated from clinical specimens; furthermore, mobile genetic elements, such as integrons and transposons, are certainly involved in their transfer.
All ESBL-positive water isolates in the confirmatory test had at least one ESBL-encoding gene amplified by PCR. Encoding genes for ESBL production are generally the same in isolates from natural environments, animals, and humans. Coque, et al. 38, highlighted the need for environmental surveillance to identify bacterial clones capable of producing ESBLs in the community and the natural environment.
Isolate Koz 6CFL from a water sample collected at Fundão Island II and isolate Kp 2141b from a different secretion revealed resistance to fourth-generation cephalosporin and to carbapenems in the AST, but the confirmatory test for carbapenem-enzyme production and PCR for blaKPC, were negative. Thus, resistance to imipenem and ertapenem seem to be associated with other resistance mechanisms. In the strains expressing resistance to fourth generation cephalosporin’s and carbapenems in the AST, we chose to evaluate only the presence of blaKPC gene and not other genes like blaOXA, blaIMP, and blaNDM. The blaKPC gene was initially detected in strains of K. pneumoniae of hospital origin, which aroused our interest to investigate the presence of this gene in environmental strains and to compare them with strains isolated from clinical samples.
Leavitt, et al. 39 identified K. neumonia strains resistant to ertapenem which were not carbapenemases producers but instead harbored genes for CTX-M, SHV, and TEM associated with changes in the cell wall permeability due to a lack of gene expression for cell-wall membrane proteins. In the present study, isolate Koz 6CFL showed the presence of genes encoding for CTX-M, SHV, and TEM while isolate Kp 2141b only presented genes encoding for CTX-M. The existence of these encoding genes in clinical isolates would have serious consequences for antibiotic therapy, and their presence in environmental isolates may indicate not-treated discharges into the natural environment which may contain antimicrobial drug residues and/or bacteria carrying resistance genes in mobile genetic elements.
Strains Ec 1Gd, Ec 4Ge, and Kp 8Gb1 isolated from the Guapimirim and Magé Rivers and from Mangue Channel, respectively, showed PCR results positive for qnrB gene. Takasu, et al. 40, detected the presence of qnrB in isolates from samples collected in bodies of water in Vietnam and concluded that the presence of this gene in such isolates was not associated with the use of quinolone antimicrobials in animal husbandry, aquaculture or infections control, although the existence of these genes in natura has not been proven to be intrinsic to the bacterial genome. According to Strahilevitz, et al. 41, the origins of qnr family genes were not only found in environmental bacteria but also in bacteria isolated from humans and animals. Nordmann, et al.42, reported that qnr genes in Enterobacteriaceae can be associated to ESBLs, CTX-M-1 or CTX-M-9, which may indicate that genes encoding for beta-lactamases and quinolone resistance mechanisms may be located in the same plasmid with a possibility of transfer to other species.
The presence in hospital settings of Enterobacteriaceae strains harboring resistance genes to several classes of antimicrobial drugs or of several genes encoding for one same class is a well-established fact 43. Besides the small bacterial population analyzed in the present study, it was possible to detect a representative group with well discernible characteristics, which is occurring in hospitals all over the world. In a study by Minarini, et al.44, Enterobacteriaceae strains harboring qnr genes were isolated from outpatients in several Brazilian states. Although the clinical isolates in this study come from inpatients, these results corroborate those reported by Minarini, et al. 44, showing that qnrB is more prevalent in Brazil than qnrA, while in the latter study, no samples from Rio de Janeiro state were studied.
Öktem, et al. 45, studying E. coli and K. pneumoniae strains isolated from blood samples showing resistance to beta-lactamic and quinolone drugs found a correlation between the plasmids containing at least one beta-lactamase encoding gene (blaTEM, blaSHV or blaCTX-M) associated with the presence of the qnrA gene. All the isolates in the study, except for Kp 2692, showed the presence of a plasmid by PCR testing. The existence of plasmids concomitantly harboring genes encoding resistance for different classes of antimicrobial drugs may promote dissemination of bacteria involved in infections of complex treatment 38.
Sewage treatment plants constitute a favorable environment for the selection and transfer of resistance genes between bacteria. Mokracka, et al. 46, studied the water from different sites at a sewage treatment plant and found E. coli strains containing integrons, even in samples from the end effluent, some of which even had genes encoding for different antimicrobial classes. In the present study, we found an isolate (Kp 2Ge) from water collected at the Caceribu River containing the aacC2, blaTEM, blaSHV, and the blaCTX-M-1 genes. There was also an isolate (Ec 4Ge) from water collected at the Magé Channel that included aacC2, blaTEM, blaCTX-M-1, and qnrB genes. Despite the fact that the Caceribu River and the Magé Channel are part of a protected area, they have been degraded due to discharges of anthropic origin, results which were corroborated by the data from the Instituto Estadual de Meio Ambiente (INEA) 19.
Among the isolates from water samples collected in the areas of higher degradation, isolates Koz 6Gb and Koz 6CFL (from Fundão Island II) contained aacC2, blaTEM, blaSHV, and blaCTX-M-1 genes. Isolates Ec 7Ga, Ec 7Gb, Ec 7Gc, and Ec 7CFLd from water samples collected at Bica beach showed the presence of aacC2, blaTEM, and blaCTX-M-1 genes, while isolate Kp8Gb from water samples collected at the Mangue Channel harbored the aacC2, blaTEM, blaCTX-M-1, and qnrB genes.
The transfer of genetic elements is occurring uninterruptedly in the hospital and the community environments either during bacterial division (vertical form) or through other mechanisms, such as bacterial conjugation (horizontal form). The selective pressure for the use of antimicrobials in these environments chooses those microorganisms that carry such elements, which will thus have the possibility of spreading to other microorganisms.
The presence of E. coli strains in water samples is an indicator of fecal contamination 47. Multidrug-resistant Enterobacteriaceae strains which colonize human and animal intestines are continuously discharged to the natural environment and may cause serious opportunistic infections behaving as reservoirs for antimicrobial-resistant genes. The genes identified in the present study indicate that the water bodies where samples were collected were exposed to sewage dumping from households, hospitals, and industries and this has prompted the presence of bacteria with the latter characteristics in these natural bodies of water.
We want to emphasize that the use of adequate procedures such as the proper use of antimicrobial drugs in hospitals, veterinary care, and aquaculture, may contribute to more efficient control of the dissemination of bacteria harboring plasmids containing genes encoding for antimicrobial-resistance mechanisms. It is clear that there is a need for a committed effort to sustain efficient microbiological monitoring and to prevent the discharges of antimicrobial drugs residues and bacteria into the natural environment. It is also paramount to detect multidrug-resistant bacteria in bodies of water given that this is an efficient way of disseminating bacteria and the traits they may carry.