Fungal pulmonary infections remain one of the most important health problems encountered in immunocompromised individuals 1. In general, mycoses became a common finding after the AIDS epidemic onset, with several fungi causing disease in this population 1. Among these fungi, Pneumocystis jirovecii (formerly known as Pneumocystis carinii), which has a worldwide distribution, causes fungal pneumonia considered an AIDS- defining disease 2. Pneumocystis jirovecii pneumonia also occurs in non- HIV-infected patients such as transplant recipients, individuals with prolonged immunosuppressive therapies, with autoimmune complications, and neoplasic disorders under chemotherapy 3.
Historically, it was believed that P. jirovecii pneumonia was a direct consequence from a latent prior infection 4, but a study of 15 lung autopsies of immunocompetent individuals aged 15-75 years showed the absence of P. jirovecii and disproved this hypothesis 5. Nowadays, the most accepted hypothesis is P. jirovecii transmission from person to person or acquisition from the environment, both by airborne route. Furthermore, disease development can also occur due to reactivation or reinfection 6. This fungus can cause an asymptomatic infection to fulminant pneumonia depending on the host’s immune status. The most severe case of this disease presents when CD4+ lymphocyte count is lower than 200 cells/ml, frequently occurring in male patients 7.
The P. jirovecii pneumonia diagnosis is difficult, mainly because P. jirovecii cannot grow in traditional mycological culture media such as Sabouraud or Mycosel 8. Cultures of P. jirovecii are obtained only in pneumocyte cell cultures, which are not available in most laboratories 9. Histopathologic diagnosis by staining methods or fungal visualization (with the fluorescent dye calcofluor white) depends on the fungal burden, specific staining and the observer’s expertise 10. In some cases, the traditional histopathologic diagnosis can be difficult, due to similar structures present in the material that can lead to a misdiagnosis with other microorganisms, making differential diagnosis strongly necessary 11. In addition, serological methods based on antibody detection have limitations in P. jirovecii pneumonia diagnosis. Humans produce antibodies against the gpA protein, used in serological studies 12. However, due to high genetic variation, this protein is not widely studied. Another serological tests limitation is that patients from different geographic regions may have different levels of immune response to this surface protein 13.
Laboratory findings are generally not helpful in the P. jirovecii pneumonia diagnosis because many findings are compatible with other infections. For example, increased serum lactate dehydrogenase is present in HIV-infected patients 14-16. Symptoms such as fever, cough, dyspnea, and, in severe cases, respiratory failure, are also manifestations of other infections, such as tuberculosis 17, histoplasmosis 18, cryptococcosis 19, community- acquired pneumonias (CAP) 20, and more recently, coronavirus disease 2019 (COVID-19) 21.
Several research groups reported the use of molecular methods to detect and diagnose P. jirovecii22-25. Molecular methods have a high sensitivity and specificity, almost without cross-reactivity, and present better results when compared with traditional diagnostic methods, including direct immunofluorescence 23 and serology 24. Up to now, nested PCR and realtime PCR are the most widely used molecular diagnostic methods, and have different levels of sensitivity and specificity, according to the protocol and the selected target 26. These methods are not able to differentiate between colonization and infection, but provide fast and accurate results, important factors because faster results imply quicker diagnosis and treatment, improving the health quality of immunocompromised populations.
Even with a multitude of available molecular tests for P. jirovecii pneumonia diagnosis, the immunofluorescence assay remains as the gold standard for the diagnosis of pneumocystosis, as it detects the presence of spore or ascus (formerly known as cysts and trophozoites) 27. However, as exposed, it is a test with several limitations and requires careful result analysis, especially in cases with low fungal load, which can easily present false negative results with this technique 23. In these cases, it is hard to interpret if a positive molecular method result (performed with the same respiratory sample) is a true false-positive or if it indicates a fungal burden below the immunofluorescence detection limit.
This study aimed to compare the results of traditional, direct immunofluorescence assay versus molecular methodologies (nested PCR and a commercial real-time PCR kit) to detect P. jirovecii in respiratory samples from immunocompromised patients with fungal pneumonia suspicion and correlate with the scoring system to predict P. jirovecii pneumonia 28.
This study was approved by the Research Ethics Committee of the Evandro Chagas National Institute of Infectious Diseases - Oswaldo Cruz Foundation (INI-Fiocruz), CAAE 00580.0.009.000-09.
Materials and methods
Patients and samples
A study was conducted (2013-2014) in patients with suspected fungal pneumonia at Instituto Nacional de Infectologia Evandro Chagas, Fundação Oswaldo Cruz, Rio de Janeiro, Brazil. Inclusion criteria were patients over 18 years old and were hospitalized. The included patients were people living with HIV, a CD4+ cell count lower than 200 cells/ml, and clinical symptoms compatible with fungal pneumonia. Patients without sufficient respiratory samples to carry out all proposed methodologies were excluded. Respiratory samples for diagnosis were collected depending on the patient’s condition and consent. Samples included spontaneous or induced sputum, tracheal wash, and bronchoalveolar lavage in a convenience sampling.
Culture
The culture was performed to check other possible fungal agents causing coinfections. After removing the airway mucus layer with citrate buffer and the N-acetylcysteine (a mucolytic agent) the samples were centrifuged at 11.500g for five minutes and cultivated in 2% Sabouraud Dextrose and Mycosel culture media (Becton Dickinson). Cultures were observed on a weekly basis during four weeks of incubation at room temperature. Possible fungi were identified by conventional mycologic techniques.
Hematoxylin and eosin, Grocott’s and calcofluor white stains
Previous treatment with cellblock fixative was done for hematoxylin and eosin, and Grocott’s stain, to yield a cellular pellet from the respiratory materials in 1.5 ml. All samples were paraffin-embedded for posterior sections measuring 3-4 µm. The sections were stained with the traditional protocol for hematoxylin and eosin to evidence samples’ cellular characteristics. Moreover, for the Grocott's method, the samples were stained with metenamine silver nitrate and counterstained with light green. The calcofluor staining was performed using a commercial reagent (Calcofluor White Stain - Fluka Inc.). The reaction was performed using one drop of the sample, one drop of 10% potassium hydroxide, and one calcofluor drop, as the manufacturer instructed.
Direct immunofluorescence
One ml of 0.1% dithiothreitol was added, as a mucolytic agent, to 1 ml of the respiratory specimens. The mixture was incubated at 37 °C for 15 minutes and the supernatant was discarded. After this step, the direct immunofluorescence assay was conducted using the Merifluor Pneumocystis kit (Meridian Biosciences, Inc, OH, USA) according to the manufacturer’s instructions. The results were observed with the fluorescence microscope Olympus-BX40® with a 400-fold magnification.
DNA extraction
The DNA was extracted from the respiratory samples using the QIAamp® DNA mini kit (Qiagen, Hilden, Germany), following all the manufacturer's recommendations. DNA concentrations were estimated using the Nanovue™ Plus Spectrophotometer (GE Healthcare, Buckinghamshire, UK).
Human housekeeping gene β-globin PCR and nested PCR for P. jirovecii
The single step polymerase chain reaction (PCR) for human housekeeping gene β-globin was performed using the primer pair βglobinF (5’-GCAAGA AAG TGC TCG GTG C-3’) and βglobinR (5’-TCA CTC AGT GTG GCA AAG GTG- 3’). The total reaction volume was 50 µl, using 10 µl of DNA, 10 mM Tris-HCI, 50 mM KCI, 2.5 mM MgCI2, 1.5 units of Taq DNA polymerase (ThermoFisher, MA, EUA), 30 pMol of each primer and 200 µM of dNTPs (ThermoFisher, MA, EUA). The PCR was conducted with the following conditions: 95 °C for 5 minutes; 45 cycles at 95 °C for 30 s, 57 °C for 30 s, 72 °C for 30 s and one final extension at 72 °C for 10 minutes 29.
The nested PCR targeting the P. jirovecii mitochondrial large subunit ribosomal RNA (mtLSUrRNA), was performed using in the first round the primers pAZ 102-H (5’- GTG TAC GTT GCA AAG TAC TC-3’) and pAZ 102-E (5’-GAT GGC TGT TTC CAA GCC CA-3’), and in the second reaction, the primers pAZ 102-X (5’-GTG AAA TAC AAA TCG GAC TAG G-3’) and pAZ 102-Y (5’-TCA CTT AAT ATT AAT TGG GGA GC-3’) 18. A total volume of 50 µl was set for each reaction, containing 10 µl DNA, 10 µM Tris-HCI, 50 µM KCI, 2.5 mM MgCI2,1.5 units of Taq DNA polymerase, 10 pM of each primer and 200 pM of dNTPs. The mix of the nested PCR was similar, except for the use of 2 µl of the first reaction, used as template for the second reaction. Both reactions were performed following the protocol previously described 13, with minor modifications: The first reaction was cycled 40 times at 94 °C for 1.5 minutes, 60 °C for 1.5 minutes and 72 °C for 2 minutes. The second round was cycled 40 times at 94 °C for 1.5 minutes, 61.7 °C for 1.5 minutes and 72 °C for 2 minutes 30.
Both reactions were performed in a Cl 000 thermalcycler (BioRad, Germany) and electrophoresed on a 1% agarose gel stained with 0.5% ethidium bromide.
Viasure real-time PCR
The VIASURE Pneumocystis jirovecii real time PCR detection kit (CERTEST BIOTEC, Spain) was designed for P. jirovecii diagnosis in respiratory samples. After DNA isolation, the identification of P. jirovecii was performed by the amplification of a mtLSUrRNA conserved region, using specific primers and a fluorescent-labelled probe (FAM). The reaction was performed according to manufacturer’s instructions in a Agilent AriaMX (Agilent Technologies, Santa Clara, USA) qPCR thermal cycler. The reactions consisted in a polymerase activation at 95 °C for 2 minutes and 45 cycles of annealing/extension at 60 °C for 50 s. The results were analized in the Aria MX software (version 1.7.1).
Pneumocystis jirovecii pneumonia diagnosis
The final diagnosis of P. jirovecii pneumonia was reached after the correlation of all test results and its classification based on the scoring system proposed by Smith, Forbes and Gazzard (1992) 28 (supplementary table 1). The response to sulphamethoxazole/trimethoprim treatment was also considered.
Table 1 Clinical-epidemiological characteristics of the 40 patients included in the study
| Variable | n | % | |
|---|---|---|---|
| Sex | |||
| Male | 30 | 75 | |
| Female | 10 | 25 | |
| Age | |||
| 18-19 | 1 | 2.5 | |
| 20-39 | 23 | 57.5 | |
| 40-59 | 16 | 40 | |
| Viral loada | |||
| Not available | 19 | 47.5 | |
| <1000 | 4 | 10 | |
| 1000-10.000 | 5 | 12.5 | |
| 10.001-100.000 | 2 | 5 | |
| >100.001 | 10 | 25 | |
| CD4+ countb | |||
| <10 | 3 | 7.5 | |
| 11-50 | 7 | 17.5 | |
| 51-100 | 17 | 42.5 | |
| 101-200 | 13 | 32.5 |
a copies/ml
b cells/ml
Statistical analysis
Concordance between the defined categories by the diagnostic methods (positive or negative) was evaluated using the kappa coefficient, interpreted as described 31. Statistical analyses were performed using GraphPad Prism (version 5.0) software. Non-parametric tests were used to compare groups of continuous variables. Categorical variables were analized using the Fisher exact test. A p-value less than 0.05 was considered significant.
Results
Patients and samples
During the study period, 40 patients fulfilled the inclusion criteria and were enrolled, resulting in 40 clinical respiratory samples (21 induced sputum, 11 tracheal wash, 5 bronchoalveolar lavage, and 3 spontaneous sputum) for analysis. Table 1 presents the clinical-epidemiological characteristics of the patients. After analyzing the clinical features of each patient, all individuals were assigned a final score to know the probability of having P. jirovecii pneumonia (table 2), ranging from 23 to 98%. All patients started sulphamethoxazole/ trimethoprim prophylaxis after the respiratory samples collection.
Staining methods and culture
Most of the samples (72.5%) showed respiratory tract cells: squamous cells, cylindrical cells, and alveolar macrophages (data not shown) while 27,5% of the samples were paucicellular or presented cell absence. Among the staining techniques performed, calcofluor white, and silver staining did not reveal compatible P. jirovecii cells in any respiratory sample. All the cultures did not demonstrate any fungal growth. The results for all tested methodologies are presented in supplementary table 2.
Direct immunofluorescence
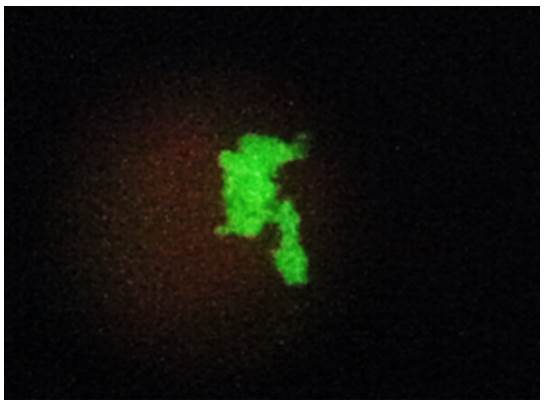
Among the 40 clinical specimens evaluated, six samples (15%) had structures morphologically compatible with P. jirovecii stained with a fluorescent green color and were considered positive by direct immunofluorescence using the Merifluor Pneumocystis kit (figure 1).
Molecular methods
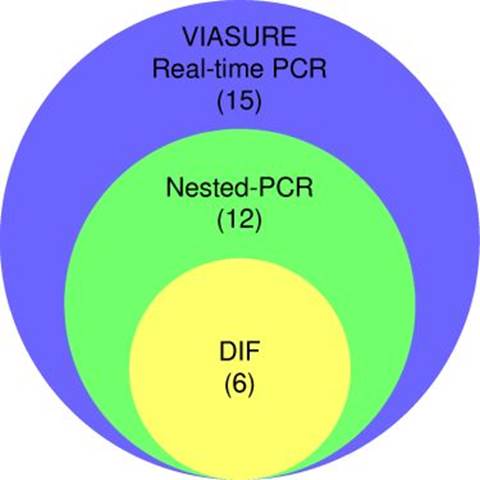
Partial amplification of the endogenous 0-globin control gene yielded a specific fragment of 79 bp, evidencing successful DNA extraction from all respiratory specimens (supplementary figure 1). Nested PCR targeting the mtLSUrRNA gene revealed 12 positive specimens (30%), with a 260 bp fragment amplification (figure 2). The VIASURE Pneumocystis jirovecii real-time PCR detection kit, designed for the same conserved region of mtLSUrRNA, amplified 15 samples (37.5%). All positive samples by direct immunofluorescence were also positive by nested PCR, and all positive samples by nested PCR were amplified by real-time PCR (figure 3).
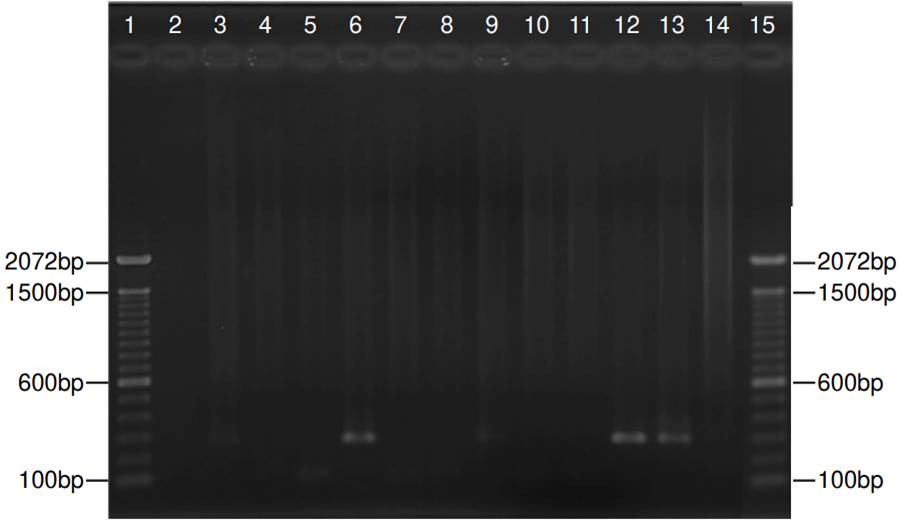
Figure 2. Representative nested polymerase chain reaction for jirovecii in 11 samples. Lanes 1 and 15: Molecular weight (1 kb plus DNA ladder - ThermoFisher Scientific, Inc.); lanes 3, 6, 9, and 12: Positive samples with a fragment of 260 bp; lanes 2, 4, 5, 7, 8,10 and 11: Negative samples; lane 13: Positive control, lane 14: Negative control. The size (base pairs - bp) of the bands is indicated in both sides.
Clinical and laboratory findings correlation
The direct immunofluorescence, nested PCR, and VIASURE real-time PCR detection kit had discordant results for P. jirovecii detection. For this reason, a clinical-laboratory correlation with the results of all methodologies was performed, and generated a scoring system for individuals with high-risk of P. jirovecii pneumonia.
The direct immunofluorescence was negative for patients with a probability of 23% (19 patients among the 40 studied) for P. jirovecii pneumonia. This technique significantly demonstrated insatisfatory results in patients with low P. jirovecii pneumonia probability (p = 0.02, Fisher’s exact test).
Nested PCR results for patients with a 23% probability of having P. jirovecii pneumonia indicated only one positive (5%) and 18 negative samples. Among patients with 82% or higher probability for P. jirovecii pneumonia, 11 (52%) samples were positive, while 10 (48%) were negative.
A statistically significant difference between these two groups (p = 0.0015, Fisher’s exact test) was detected. The kappa index demonstrated a moderated correlation (0.583) between direct immunofluorescence and nested PCR (figure 3).
The results of VIASURE Pneumocystis jirovecii real-time PCR evidenced a statistically significant difference between these two groups (p = 0.0011, Fisher’s exact test); firstly, the group with 23% of PCP and in the second, the group higher than 82% of PCP. The kappa index demonstrated a moderated correlation (0.455) between direct immunofluorescence and real-time PCR results. In addition, an agreement was observed between nested PCR and real-time PCR (kappa = 0.833). All these results are summarized in table 3 and supplementary table 2.
Table 3. Positive results in the different Pneumocystis jirovecii diagnostic methods among four possible scores of high-risk individuals to Pneumocystis jirovecii pneumonia
| P. jirovecii pneumonia probability (n) | DIF | Nested PCR | VIASURE Real-time PCR | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| 98% 11 | 2 | 18 | 6 | 54 | 7 | 63 |
| 87% 2 | 1 | 50 | 1 | 50 | 1 | 50 |
| 82% 8 | 3 | 37.5 | 4 | 50 | 5 | 62.5 |
| 23% 19 | 0 | 0 | 1 | 5 | 2 | 10 |
DIF: Direct immunofluorescence
Discussion
This study evaluated the performance of methods for the diagnosis of pneumonia caused by P. jirovecii. The analyzed samples consisted of respiratory material collected from 40 patients with AIDS, with considerable immunosuppression and respiratory symptoms compatible with those presented by fungal diseases. The use of the scoring system proposed by Smith, Forbes and Gazzard (1992) in the included patients, allowed us to predict different probabilities of P. jirovecci pneumonia among them.
The gold standard method to detect P. jirovecii is direct immunofluorescence, since this technique detects P. jirovecii in its two lifecycle stages 32. Conventional techniques have shown intrinsical limitations, such as microscopic visualization of clinical samples which depends on cellular quality, microorganism amount, and observer expertise 26. In our sampling, the gold standard method showed the lowest positivity among the other diagnostic techniques tested. Direct immunofluorescence makes it possible to detect P. jirovecii only in patients with a higher probability of P. jirovecii pneumonia. It may indicate a failure of P. jirovecii pneumonia diagnosis in oligosymptomatic patients or at early stages of the disease. Our results corroborate the indication of other research groups about the use of molecular methods as alternative techniques to detect this fungal pathogen.
Several reports have shown the low sensitivity of conventional techniques when compared to molecular methods. A study with 275 clinical respiratory specimens, comparing Grocott's stain, direct immunofluorescence, and nested PCR (targeting the mtLSUrRNA) revealed nine positive samples by the Grocott's stain and 16 positive samples by direct immunofluorescence. However, when the samples were evaluated by nested PCR, the authors found P. jirovecii DNA in 44 samples 26.
Another report, using 50 clinical specimens of respiratory origin and comparing three methodologies: Giemsa, direct immunofluorescence, and nested PCR (targeting the mtLSUrRNA), showed that Giemsa stain detected P. jirovecii only in one sample, direct immunofluorescence yielded four positive results and nested PCR resulted in 36 positive samples 33.
A study comparing nine molecular methods showed that nested PCR targeting the mtLSUrRNA gene should be considered the most sensitive technique 34 for P. jirovecii pneumonia diagnosis. The explanation for the better performance of this technique is that the mtLSUrRNA gene is fully involved in basic metabolic processes, and it has a high degree of genetic conservation 35 in P. jirovecii. In addition, the high number of P. jirovecii mitochondrias makes the mtLSUrRNA gene an excellent target for P. jirovecii pneumonia diagnosis 26, increasing the sensitivity of the nested PCR. For this reason, this partial sequence has become the molecular target most suitable for P. jirovecii detection. The results herein corroborate the utility of the mtLSUrRNA gene for early detection of pneumonia caused by P. jirovecii36.
In contrast with most eukaryotic organisms, P. jirovecii has only one copy of the gene encoding the ribosomal RNA, and this explains why techniques that use internal transcribed spacer (ITS) region as diagnostic target has low efficiency 37. The VIASURE real-time PCR kit was also developed to target the mtLSUrRNA gene. The results obtained in this study were better than those of nested PCR, with three additional patients diagnosed with P. jirovecii pneumonia. These results suggest that the VIASURE real-time PCR should be used for P. jirovecii pneumonia diagnosis in laboratories with the structure to perform it.
The lack of studies with this commercial kit does not allow us to establish a difference between P. jirovecii colonization and infection. However, our results together with the proposed clinical score indicated a possible P. jirovecii infection in the evaluated patients. To the best of our knowledge, this is the first study associating the Smith et al. (1992) score with laboratory data. Positive samples were detected in patients with lower P. jirovecii pneumonia probabilities based on the scoring system, which leads us to believe that this detection occurred in the early stages of the disease. Among the 40 studied patients, we observed that nested PCR was positive in the 12 samples, and among these, six were also positive by direct immunofluorescence. The increase in the positivity rate associated with the scoring system made us conclude that molecular methods, although not new, remain useful tools for the P. jirovecii pneumonia diagnosis.
Nested-PCR is an accessible technique for many laboratories, and the results are not observer-dependent like those of direct immunofluorescence. However, cross-contamination may occur between the first and second reactions, so laboratories should be careful when handling these samples. Real-time PCR provides fast results, also observer-independent, and have a low chance of external DNA contamination. However, it requires expensive equipment that may not be available in all centers handling patients with AIDS.
With the results of this work, it was concluded that nested PCR provides accurate results in laboratory practice for pneumocystosis diagnosis, especially in laboratories with basic infrastructure. However, the VIASURE real-time PCR showed superior results compared to nested PCR. Therefore, we suggest that, in countries where the kit is approved for diagnostic use, it should be performed as a routine test when the necessary infrastructure is available. Molecular results should be interpreted along with each patient’s clinical signs and symptoms, and the score previously suggested 28 is adequate for this purpose.