Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Acta Biológica Colombiana
Print version ISSN 0120-548X
Acta biol.Colomb. vol.14 no.3 Bogotá Sep./Dec. 2009
GENETIC VARIABILITY OF THE ZEBU CATTLE BREED (Bos indicus) IN THE DEPARTAMENT OF HUILA, COLOMBIA USING MICROSATELLITE MOLECULAR MARKERS
Variabilidad genética de Zebú (Bos indicus) en el departamento del Huila, Colombia, usando marcadores moleculares por microsatélites
CARLOS HERNÁNDEZ ESCOBAR1,2*,MVZ, M.Sc (C); MARTHA OLIVERA ÁNGEL2, MV, Dr sci Agr; HENRY OSTOS ALFONSO1, M.D., M.Sc; MARIA TERESA GUERRA1, Biol.
¹ Laboratorio de Medicina Genómica, Facultad de Ciencias de la Salud, Universidad Surcolombiana,
² Grupo Biogénesis, Facultad de Ciencias Agrarias, Universidad de Antioquia. AA 1226, Medellín, Colombia. caherez@yahoo.es*
Received 11 September 2008, Revised 6 August 2009 - Acepted 17 September 2009
ABSTRACT
The polymorphism of 11 microsatellites from zebu cattle (Bos indicus) was studied using a commercial multiplex system to estimate genetic variability. Allele frequencies polymorphism information content and heterozygosis were calculated. Allele frequencies revealed that in the analyzed sample the markers were not equally polymorphic. The average allele was 14.2 with the highest values for the TGLA122 microsatellites. The mean heterozygocity was 0.7056 and the polymorphism information content was 0.668. This multiplex analysis could be used for pedigree information and for adequate genetic improvements in breeding programs and paternity test.
Key words: alleles, microsatellite, polymorphism, zebu.
RESUMEN
Para estimar la variabilidad genética, el polimosfismo de 11 microsatélites de vacunos zebú (Bos indicus) fue estudiado mediante el sistema comercial multiplex system. Se calcularon frecuencias alélicas, contenido de información polimórfica y heterocigosis. Las frecuencias alélicas de la muestra analizada revelan que los marcadores no fueron equitativamente polimórficos. El alelo promedio fue 14,2 con el mayor valor para los microsatélites TGLA122. El promedio de heterocigosidad fue 0,7056 y el contenido de información polimórfica de 0,668. Este tipo de análisis puede ser usado para información de pedigrí y mejoramiento genético en programas de cría y pruebas de paternidad.
Palabras clave: alelos, microsatélite, polimosfismo, zebú.
INTRODUCTION
The Bos indicus Zebu herd with the Brahman (Red and White) and Gyr breeds is one of the largest commercial beef herds in the world and is well-adapted to tropical regions. The Zebu cattle breed is the most important beef herd in Colombia, where the total number of purebred and crossbreed Zebu cattle totals 95% of the bovine population. Accurate pedigree information is essential to maintain the quality of breed improvement programs and molecular markers have become an important genetic tool in genetics studies, allowing the analysis of genetic variability within and between herds. Microsatellites have been widely used as genetic markers in bovine population studies and pedigree verification (Visscher et ál., 2002; Hansen et ál., 2002; Ibeagha-Awemu and Erhardt, 2005), mainly because of their large polymorphism information content, widespread distribution in the eukaryotic genome (Tautz and Renz, 1984) and the robust methodology available. Microsatellites have been effective in evaluating differences within cattle breeds and in determining population substructures (MacHugh et ál., 1998; Ciampolini et ál., 1995). More than 1400 microsatellites have been mapped in the cattle genome (Luikart et ál., 1999) and some of them have been employed in population genetics studies and kinship verification. The aim of the study described in this paper was to characterize Zebu cattle through the analysis of the genetic variability of eleven microsatellite markers and to evaluate if these markers provide information on the genetic variability and parentage test of this herd. This study presents preliminary information to determine the possible use of microsatellite molecular markers in evaluating parentage test in a zebu cattle population (Bos indicus) in the Huila region.
MATERIALS AND METHODS
SAMPLE COLLECTION AND DNA EXTRACTION
Eighty unrelated adult Zebu cattle (47 Brahman and 33 Gyr breeds) were sampled. They were registered in their breeding associations and randomly selected from private and research herds belonging to 7 farms located in different regions of department Huila (Colombia). Blood samples were collected in EDTA with tubes and total genomic DNA was isolated by the Salting out method (Aljanabi and Martinez, 1997) and stored at -20 °C.
MICROSATELLITE AMPLIFICATION
As recommended by the International Society of Animal Genetics (ISAG), eleven microsatellites (Table 1) were selected for the analyses using the Stockmarks for Cattle Bovine Genotyping Kit (Applied Biosystems, Division Perkin-Elmer, Foster City, CA). Multiplex amplification was carried out in a final volume of 15 µL containing 50 ng template DNA, 0.5 units AmpliTaq GoldTM polymerase (PE Applied Biosystems, Foster City, CA), 3.0 µL Stockmarks Buffer, 400 µM each dNTP and 5.5 µL primer mix (Table 1). A Programmable Thermal Controller PTC-100 (MJ Research, INC) was used in an initial denaturation phase of 15 min at 95 °C, followed by 31 cycles of 45 s at 94 °C, 45 s at 61 °C and 1 min at 72 °C. A final extension was programed at 72 °C for 1 h and then at 25 °C for 2 h. After amplification, 90 µL water were added to each tube and 0.4 µL of this solution was mixed with 2 µL loading mix (DI formamide: dye: GS 500 Rox -6:1:1) and analyzed using an ABI PRISM 3100 DNA Sequencer. The fluorescence data wascollected by GeneScanTMAnalysis 3.0 and analyzed using GenotyperTM3.0 software.Parentage testing was carried out by assessing compatibility between alleles present ina calf and those found in the assumed parents. As suggested by Luikart et ál., 1999 andWeller et ál., 2004, an assigned parent was excluded if its genotype was incompatible intwo or more loci with that of the offspring, but parentage was not excluded if incom-patibility occurred in only one locus.
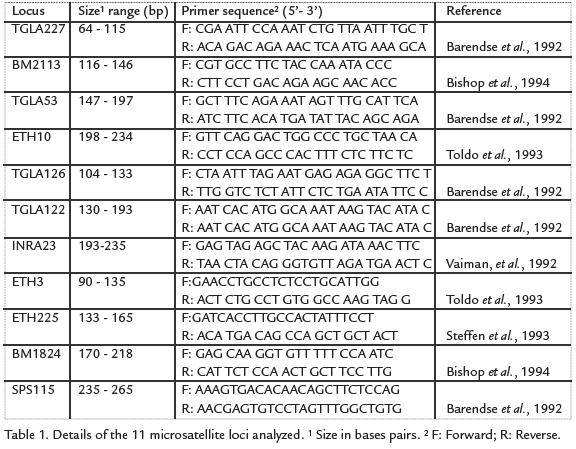
DATA ANALYSIS
The ARLEQUIN package Version 3.1 (Schneider et ál., 2005) was used to calculate anexact test for deviation from Hardy-Weinberg equilibrium (HWE), allele and geneticsfrequencies, heterozygotic deficiency, expected heterozygosity (He) and observedheterozygosity (Ho), and polymorphism information content (PIC).
RESULTS
One hundred and fifty seven alleles were detected from the 11 loci surveyed, yielding amean value of 14.2 alleles per locus. The allele frecuencies of 11 microsatellites are listedin Table 2. Allele frequencies revealed that not all markers were equally informative. Thenumber of alleles per locus ranged from nine for ETH10 to 17 for BM2113. Tree loci(TGLA227, INRA23 and ETH3) deviated significantly (p < 0.05) from the HWE. A significant deficit of heterozygosity (p < 0.01) was detected in the BM2113, TGLA 53,ETH10, SPS115, TGLA126, TGLA122, ETH225 and BM1824 loci. The mean PIC valuewas 0.668 and the mean expected heterozygosity value was 0.773. The expected andobserved heterozygosity range from 0.411 for TGLA227 to 0.876 for TGLA122 andfrom 0,334 for BM1824 to 0.785 for ETH3 respectively, the TGLA122 locus showedthe highest allele polymorphism, values are shown in Table 3.
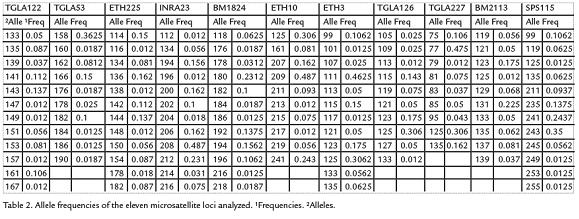
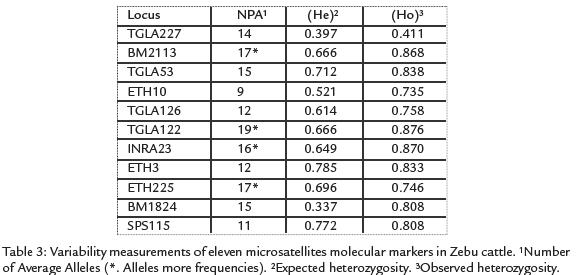
DISCUSSION
Accurate cattle pedigree information is essential for optimal development of breed andselection programs to improve productivity in farm animals. Misidentification ofparentage can lead to breeding inaccuracy, causing great financial losses in the beefindustry (Geldermann et ál., 1986). Microsatellites are the most widely used molecularmarkers in pedigree control. The use of microsatellites with high polymorphism in-formation content would help to correctly identify individual cattle to improve cattlebreeding programs. Cervini et ál., 2006, identified the genetic variability of BrazilianNelore cattle population, detecting that the markers TGLA 227, BM1824 and TGLA53loci each had one allele with a much higher frequency than the other allele (75 bp, 180bp, 160 bp respectively). The number of allele per locus ranged from six for TGLA227to 16 for TGLA122. The TGLA 122 locus showed the highest allele polymorphism, whilethe INRA023 locus displayed the highest exclusion probability. The mean PIC value was0.640 and the mean expected heterozygosis value was 0.679.
Little information is available regarding the allele frequencies of the eleven microsatellitesstudied here as well as for other variability estimates for Brahman cattle. Furthermore, todate in Colombia no estimates exist of the multiplex variability in cattle. Since the evaluation of polymorphism depends strictly on allele numbers and their frequency distribution,estimates of allele frequencies are essential. A comparison of the results obtained for B.indicusZebu cattle with those of B. taurusbreeds (Peelman et ál., 1998; Heyen et ál., 1997)indicated a difference in variability for some loci, which are highly informative in B. taurusbut less informative in B. indicus. Ten microsatellite loci assess the feasibility of applying in parentage control of beef cattle in Portugal, present in total number of alleles found for the 10 microsatellite markers was 107, the overall of loci mean (na) per locus was 8.20, while it was for TGLA53: 10.67 and for TGLA227: 10.00 loci, while the lowest mean was observed for BM1824 (5.67). The mean He for the set of 10 microsatellites used was 0.733, all the loci showed high levels of genetic variability, with heterozygosity ranging between 0.587 (ETH10) and 0.837 (BM2113) and in general microsatellites BM2113 and TGLA53 were the most informative loci (Carolino et ál., 2009).
According to Peelman et ál., 1998, who studied Belgian cattle, the number of TGLA53 locus alleles in Holstein Friesian (13 alleles), Belgian Red Pied (12 alleles), East Flemish (12 alleles) and Belgian Blue (10 alleles) cattle were very similar to those found in Nellore cattle (13 alleles). However, we found that the exclusion probability for the TGLA53 locus in Brazilian Nellore (EP = 0.256) cattle is much lower than in the four Belgian breeds (Holstein Friesian = 0.742, Belgian Red Pied = 0.711, East Flemish = 0.698 and Belgian Blue = 0.682). We obtained similar results for the TGLA227 locus (EP 0.230), much lower values than those described by Heyen et ál., 1997, for Holstein (0.69), Red Angus (0.63) and Gelbvieh (0.68) cattle. Thus, the efficiency of these markers in European B. taurus cattle is not always the same as for Indian B. indicus zebu (Brahman, Nellore).
In the present study we found significant (p< 0.01) deviations from HWE for six loci (TGLA122, INRA023, TGLA53, ETH10, ETH225 and ETH3). Machado et ál., 2003, also reported significant deviations from HWE for Brahman and Gyr cattle breeds using microsatellite markers. Almeida et ál., 2000, concluded that the TGLA122 locus was in HWE in the Brazilian hybrid bovine breed. We observed deviations from HWE caused by heterozygote deficiency at the TGLA122, INRA023, TGLA53, ETH10 and ETH225 loci. Beja-Pereira et ál., 2003 and Loftus et ál., 1999, reported deviations from HWE in other European bovine populations, also caused by heterozygosity deficit, and similar results have been reported by Loftus, 1999, in six populations, including Indian Zebu cattle.
Several factors can lead to heterozygote deficiency including null alleles, assortive mating, the Wahlund effect, selection against heterozygotes, inbreeding, or a combination of these factors. Null alleles are alleles that are not amplified (usually due to a mutation in one of the primer binding sites) and are commonly reported in microsatellite studies as being the source of heterozygosity deficit (Pemberton et ál., 1995). The frequency of microsatellite loci containing null alleles has proved to be as high as 30% in humans (Callenet ál., 1993). In paternity tests, an undetected null allele may have profound consequences, since it may cause rejection of an otherwise correctly assigned parent (Holm et ál., 2001).
To date, there are no reports of studies on Zebu cattle indicating the presence of null alleles for the markers analyzed, although the presence of null alleles has previously been observed in segregation analyses using other microsatellite loci in Zebu cattle (Tambasco et ál., 2000). This possibility cannot be excluded because segregation analysis using the loci evaluated in this study has not yet been undertaken for Zebu cattle. Despite the paucity of information provided by some of the loci analyzed here, the use of this multiplex analysis proved efficient in Zebu characterization and can be used for pedigree verification.
The genetic information like heterozygosity, Hardy Weinberg Equilibrium and allele frequencies allowed relations between this parameter with the inbreeding and other traits productive and evolutionary with high association in the population structure evaluated, the genetic association with the results allows us to find that the level of variability is directly linked to inbreeding and diversity low, affecting the potential production of different breeds.
ACKNOWLEDGEMENTS
We thank the private farm association for sample selection. This study was made possible by the Governing of Huila, Colombia, and the Science and Technology Department Council CODECYT by financial support.
REFERENCES
ALJANABI SM, MARTINEZ I. Universal and rapid salt extraction of high quality genomic DNA for PCR-based techniques. Nucl Acids Res. 1997;25(22):4692-4693. [ Links ]
ALMEIDA EA, WEIMER TA. Genetic Diversity of a Brazilian Crerole cattle Based on Fourteen Microsatellite Loci. Arch Zootec. 2004;53:3-11. [ Links ]
BARENDSE W, ARMITAGE SM, KOSSAREK LM, SHALOM A, KIRKPATRICK BW, et ál. A genetic linkage map of the bovine genome. Nat Genet. 1994; 6:227-235. [ Links ]
BEJA-PEREIRA A, ALEXANDRINO P, BESSA I, CARRETERO Y, DUNNER S, et ál. Genetic characterization of Southwestern European bovine breeds: A historical and biogeographical reassessment with a set of 16 microsatellites. J Hered. 2003;94:243-250. [ Links ]
BISHOP MD, KAPPES SM, KEELE JW, et ál. A genetic linkage map for cattle. Genetics. 1994;136:619-639. [ Links ]
CALLEN DF, THOMPSON AD, SHEN Y, PHILLIPS HA, RICHARDS RI, et ál.. Incidence and origin of null alleles in the (AC)n microsatellite markers. Am J Human Genet. 1993;52:922-27. [ Links ]
CAROLINO INÊS, CONCEIÇÃO O. et ál.. Implementation of a parentage control system in Portuguese beef-cattle with a panel of microsatellite markers. Genet Mol Biol. 2009;32(2)306-311. [ Links ]
CERVINI M, HENRIQUE-SILVA F, MORTARI N, MATHEUCCI E Jr. Genetic variability of 10 microsatellite markers in the characterization of Brazilian Nellore cattle (Bos indicus). Genet Mol Biol. 2006;29(3)486-490. [ Links ]
CIAMPOLINI R, MOAZAMI-GOUDARZI K, VAIMAN D, DILLMAN C, MAZZANTI E, et ál.. Individual multilocus genotypes using microsatellite polymorphisms to permit the analysis of the genetic variability within and between Italian beef cattle breeds. J Animal Sci. 1995;73:3259-68. [ Links ]
GELDERMANN H, PIEPER U, WEBER WE. Effect of misidentification on the estimation of breeding value and heritability in cattle. J Animal Sci. 1986;63:1759-68. [ Links ]
HANSEN C, SHRESTHA JN, PARKER RJ, CROW GH, MCALPINE PJ, DERR JN. Genetic diversity among Canadienne, Brown Swiss, Holstein, and Jersey cattle of Canada based on 15 bovine microsatellite markers. Genome 2002;45:897-904. [ Links ]
HEYEN DW, BEEVER JE, DA Y, EVERT RE, GREEN C, et ál.. Exclusion probabilities of 22 bovine microsatellite markers in fluorescsnt multiplexes for semi-automated parentage testing. Animal Genet. 1997;8:21-27. [ Links ]
HOLM LE, LOESCHCKE V, BENDIXEN C. Elucidation of the molecular basis of a null allele in a rainbow trout microsatellite. Marine Biotechnol. 2001;3:555-60. [ Links ]
IBEAGHA-AWEMU EM, ERHARDT G. Genetic structure and differentiation of 12 African Bos indicus and Bos taurus cattle breeds, inferred from protein and microsatellite polymorphisms. J Anim Breed Genet. 2005;122:12-20. [ Links ]
LOFTUS R, ERTRUGRUL O, HARBA A, EL-BARODYS M, MACHUGH D, et ál. A microsatellite survey of cattle from a centre of origin: The Near East. Mol Ecol. 1999;8:2015-2022. [ Links ]
LUIKART G, BIJU-DUVAL M-P, ERTUGRUL O, ZAGDSUREN Y, MAUDET C, TABERLET P. Power of 22 microsatellite markers in fluorescent multiplexes for parentage testing in goats (Capra hircus). Animal Genet. 1999;30:431-438. [ Links ]
MACHADO M, MARTINEZ M, et ál.. Genome scan for QTLs related to tick resistance in bovine. Genet Mol Biol. 2003;23:379-384. [ Links ]
MACHUGH DE, LOFTUS RT, CUNNINGHAM P AND BRADLEY DG. Genetic structure of seven European cattle breeds assessed using 20 microsatellite markers. Animal Genet. 1998;29:333-40. [ Links ]
PEELMAN LJ, MORTIAUX F, VAN ZEVEREN A, DANSERCOER A, MOMMENS G, et ál.. Evaluation of the genetic variability of 23 bovine microsatellite markers in four Belgian cattle breeds. Animal Genet. 1998;29:161-67. [ Links ]
PEMBERTON JM, SLATE J, BANCROFT DR, BARRET JA. Nonamplifying alleles at microsatellite loci: A caution for parentage and population studies. Mol Ecol. 1995;4:249-52. [ Links ]
SCHNEIDER S, ROESSLI D, EXCOFFIER L. Arlequin version 3.1: a software for population genetics data analysis. Geneva, Switzerland, Genetics and Biometry Laboratory, University of Geneva; 2005. [ Links ]
TAUTZ D, RENZ M. Simple sequences are ubiquitous components of eukaryotics genomes. Nucleic Acids Res 1984;12:4127-38. [ Links ]
VISSCHER PM, WOOLLIAMS JA, SMITH D, WILLIAMS JL. Estimation of pedigree errors in the UK dairy population using microsatellite markers and the impact on selection. J Dairy Sci. 2002;85:2368-75. [ Links ]
WELLER JI, FELDMESSER E, GOLIK M, TAGER-COHEN I, DOMOCHOVSKY R, et ál. Factors affecting incorrect paternity assignment in the Israeli Holstein population. J Dairy Sci. 2004;87:2627-2640. [ Links ]














