Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Acta Biológica Colombiana
Print version ISSN 0120-548X
Acta biol.Colomb. vol.20 no.1 Bogotá Jan./Apr. 2015
https://doi.org/10.15446/abc.v20n1.41946
Artículo de investigación
GENETIC STATUS, SOURCE AND ESTABLISHMENT RISK OF THE GIANT TIGER SHRIMP (PENAEIDAE: Penaeus monodon), AN INVASIVE SPECIES IN COLOMBIAN CARIBBEAN WATERS
Estado genético, origen y riesgo de establecimiento del camarón tigre gigante (Penaeidae: Penaeus monodon), una especie inavasora en aguas del Caribe colombiano
J.C. AGUIRRE-PABÓN 1, G. Jr. OROZCO BERDUGO 1, J.C. NARVÁEZ BARANDICA1
1 Laboratorio de Genética Molecular, Universidad del Magdalena. Carrera 32 n.#º 22-08, sector San Pedro Alejandrino. Santa Marta, Colombia.
For correspondence. jumantis@gmail.com
Received 6th February 2014, Returned for revision 22nd April 2014, Accepted 19th May 2014.
Citation / Citar este artículo como: Aguirre-Pabón J.C., Orozco Berdugo G. Jr., Narváez Barandica J.C. Genetic Status, Source and Establishment Risk of the Giant Tiger Shrimp (Penaeidae: Penaeus monodon), an Invasive Species in Colombian Caribbean Waters. Acta biol. Colomb. 2015;20(1):117-127. doi: http://dx.doi.org/10.15446/abc.v20n1.41946.
ABSTRACT
The tiger shrimp (Penaeus monodon) is an Indo-Pacific species. Its global production between 1970 and 1980 exceeded all other shrimp species, which favored its introduction and cultivation outside its natural range in several countries of Africa, Europe, USA and South America. It is currently found in the coast of the Atlantic Ocean (Mexico, United States, Puerto Rico, Brazil, Guyana, Venezuela and Colombia). Despite the risk involved, no studies have been conducted to evaluate their impact as a possible invasive species and their genetic condition. This study evaluated the genetic status and population origin of P. monodon in the northernmost Colombian Caribbean, analyzing the mitochondrial DNA control region (mtDNA-CR). 16 individuals were randomly collected from Golfo de Salamanca and 342 original Indo-Pacific sequences were obtained from GenBank. Parameters of genetic diversity and genetic relationships were analyzed. These results were a total of 358 sequences compared and 303 haplotypes identified. Three haplotypes were identified in the Colombian population. This results showed lower genetic diversity compared with Indo-Pacific populations. These haplotypes were closely related to those found in samples from the Philippines and Taiwan. We discuss the need to create a regional network to characterize the established populations in the Great Caribbean, with the purpose of inferring colonization processes and the establishment of management measures.
Keywords: biological invasion, Caribbean Sea, founder effect, genetic relationships, mitochondrial DNA.
RESUMEN
El camarón tigre (Penaeus monodon) es una especie del Indo-Pacífico. Su producción mundial entre 1970 y 1980 superó todas las otras especies de camarón, lo que favoreció su introducción y cultivo fuera del área de distribución natural en varios países de África, Europa, EE.UU. y América del Sur. Actualmente se encuentra en la costa del Océano Atlántico (México, Estados Unidos, Puerto Rico, Brasil, Guyana, Venezuela y Colombia). A pesar del riesgo que implica, no se han realizado estudios para evaluar su impacto como posible especie invasora y su condición genética. Este estudio evaluó el estado genético y el origen de la población de P. monodon en el norte del Caribe colombiano, analizando la región control del ADN mitocondrial (ADNmt-CR). 16 individuos fueron recolectados al azar del Golfo de Salamanca y 342 secuencias originales de muestras del Indo- Pacífico fueron obtenidas de GenBank. Se analizaron los parámetros de diversidad genética y las relaciones genéticas. Se analizaron un total de 358 secuencias y se identificaron 303 haplotipos. En la población de Colombia se identificaron tres haplotipos, mostrando una baja diversidad genética en comparación con las poblaciones del Indo-Pacífico. Estos haplotipos se encontraron cercanamente relacionados con secuencias obtenidas de muestras de Filipinas y Taiwán, principalmente. Se discute la necesidad de crear una red regional para caracterizar las poblaciones establecidas en el Gran Caribe, con el propósito de inferir los procesos de colonización y el establecimiento de medidas de manejo.
Palabras clave: ADN mitocondrial, efecto fundador, invasión biológica, Mar Caribe, relaciones genéticas.
INTRODUCTION
The giant tiger shrimp (Penaeus monodon) is a native species of the Indo-Pacific Ocean, distributed in East Africa (Tanzania, South Africa, Mozambique and Madagascar) throughout the Arabian Peninsula, Southeast Asia (India, Thailand, Vietnam, China, Korea) including the islands of Taiwan, Japan, Philippines, Indonesia, Madagascar, Fiji and Australia. Since P. monodon is subject to intense exploitation in the wild and is the most farmed species in aquaculture, it became the most commercially important shrimp species, worldwide (Hulata, 2001). During the 1970s and 80s, global production of tiger shrimp exceeded that of all other shrimp species, favored widely by its introduction and farming outside their native range in various West Africa countries (Senegal, Gambia, Ivory Coast, Guinea and Angola) Europe (Italy and Cyprus), Hawaii, United States and South America (Colombia, Venezuela, Ecuador, Peru and Brazil).
The introduction of this species for cultivation brought about the invasion of native ecosystems in several countries around the world, including some in the western Atlantic. Several hypotheses have been proposed to explain the introduction of P. monodon in the Caribbean ecosystems, including: 1. Individuals escaped from aquaculture facilities into the surrounding environment as a result of accidental release during harvesting as well as mass escape during flooding events during storms and hurricanes, 2. Migrations from areas where tiger shrimp have previously become established in the wild, and 3. Through ballast water discharge, although this latter hypothesis is less feasible. Some cases of farmed shrimp that have escaped and become established in the natural environment are known, for example, in several countries of West Africa and Hawaii resulting in relatively common catches, suggesting establishment of populations in the natural environment (CABI-ISC; http://www.cabi.org/isc/datasheet/71093). In the United States, these shrimp have been captured from the coasts of North Carolina to Texas, after an accidental escape of roughly 2000 animals from an aquaculture facility in South Carolina in 1988. However, it is not believed that these animals established new population, catches have increased, starting with six animals in 2006 and increasing to 273 in 2011 (Knott et al., 2012).
The situation in the coasts of South America is similar. Fishermen on the Atlantic coast from Guyana to Colombia and off the coast the Dominican Republic in the Caribbean Sea occasionally report catches of giant shrimp. The first official reports began in Brazil in the States of Maranhao (Fausto-Filho, 1987; Santos and Coelho, 2002), Pernambuco, Alagoas (Coelho et al., 2001) Amapa (Silva et al., 2002) and Sergipe (Santos and Coelho, 2002). In Venezuela it has been reported officially in the Anzoátegui coast, in the Orinoco River delta in Golfo de Paria (Aguado and Sayegh, 2007; Altuve et al., 2008). In Puerto Rico they have been found in the Boquilla de Mayagüez (Ramos, 2012). Their catch in Colombia has been recorded more often in the Caribbean, mainly in the Golfo de Morrosquillo (Álvarez-León, 1982) and Bahía de Cispatá in Córdoba Province (Álvarez-León and Gutiérrez-Bonilla, 2007); the Cabo de la Vela and Punta Gallinas in Guajira Province (Gómez-Lemos and Campos, 2008); and the Ciénaga Grande de Santa Marta in Magdalena Province (SIBM-INVEMAR, 2008).
In populations, introduced and established through a founder effect, the ability to adapt to a new environment depends not only on external factors such as predation, competition, diseases or characteristics of the habitat invaded, but also on their genetic potential (Holland, 2000; Tsutsui and Case, 2001) because high genetic diversity is positively correlated with invasive success (Williamson, 1996). Thus, a large number of introduced individuals with high genetic diversity are predicted to increase the risk of establishment, spread, and adaptation to new habitats (Garcia-Ramos and Rodriguez, 2002; Stepien et al., 2002; Lockwood et al., 2005; Stepien et al., 2005). Even if a founder effect is caused by temporal and spatial waves of introductions originating from multiple founding sources, it may fuel the genetic diversity of an invasion, enhancing the capacity to adapt to new and changing environments (Stepien et al., 2005). Invasions caused by a small number of individuals usually exhibit low genetic diversity due to the limited gene pool when compared to source populations and this may cause demographic bottlenecks as an effect in the post-introduction phase. In these populations genetic drift and inbreeding have a greater impact and natural selection is less effective (Frankham et al., 2002; Allendorf and Luikart, 2007), resulting in a reduction in survival and reproductive success of individuals (Frankham et al., 2002), with a consequent loss of the genetic diversity and adaptive potential of the population.
Despite the threat that P. monodon represents for native biodiversity and ecosystem stability in the Great Caribbean, because of its predatory habits, high fertility rate, it carries a variety of exotic pathogens (e.g. virus) and apparently does not have any natural predators, research on its economic and ecological impact is currently lacking. Likewise, nothing is known about their origin and the genetic potential to establish, proliferate and colonize new environments. Therefore, the aim of this study was to determine the origin of invasive populations of Penaeus monodon in the Colombian Caribbean and assess their genetic diversity for the purpose of discussing about the risk of establishment in local environment. In this study we used the mitochondrial DNA control region due to its smaller effective population size, more rapid extinction of lineages, and lack of recombination would be especially useful in detecting low genetic variability in new populations from founder effects (Avise, 2000).
MATERIALS AND METHODS
Collection of samples and DNA isolation
Tissue samples from 16 specimens of P. monodon were collected in the Golfo de Salamanca, Magdalena Province, Colombia (11°2'20" N-74°15'11" W) during April 2012, (Fig. 1). Tissues of pleopods were preserved in absolute alcohol and DNA was isolated using a commercial kit (DNA Extraction MasterPure™ of Epicentre Biotechnologies, U.S.A.).
Amplification and sequencing
The mitochondrial DNA control region (mtDNA-CR) was amplified by PCR using primers 12S (forward) 5'-AAGAACCAGCTAGGATAAAACTTT-3'and PCR-1R (reverse), 5'-GATCAAAGAACATTCTTTAACTAC-3' from Chu et al., (2003). The reaction was done in a 25 μL reaction volume with a final concentration of 1x Buffer, 2 mM MgCl2, 0.2 mM of dNTPs, 0.3 µM of primers, 0.05 U of Taq DNA Polymerase and 50–100 ng/mL of template DNA. The PCR was run in a thermocycler ESCO Swift ™ MaxPro with the following temperature profiles: initial denaturation at 94 °C for 5 min followed by 30 cycles of 94 °C for 1 min, 57.2 °C for 30 s for alignment, and 72 °C for 1 min for extension, and a final extension at 72 °C for 3 min. The amplified products were purified and sequenced at Macrogen Inc. Additionally, for phylogeogrpahic comparisons, 342 sequences from the Indo-Pacific populations (Ky: Kenya, Mg: Madagascar, Ind: India, ThE: Thailand East, ThW: Thailand West, Vn: Vietnam, Ch: China, TaE: Taiwan East, TaNE: Taiwan Northeast, TaW: Taiwan West, TaS: Taiwan South, Ph: Philippines and Aus: Australia) were obtained of GenBank with accession numbers EU426576 – EU426831, FJ226007 – FJ226027, EU368046 – EU368113 and DQ311142.
Analysis of information
All sequences were edited and aligned in the programs BioEdit v7.0.5 (Hall, 2001) and MEGA v5.02 (Tamura et al., 2011). The alignment quality was also inspected visually.
Genetic status of the invasive species P. monodon in Colombia
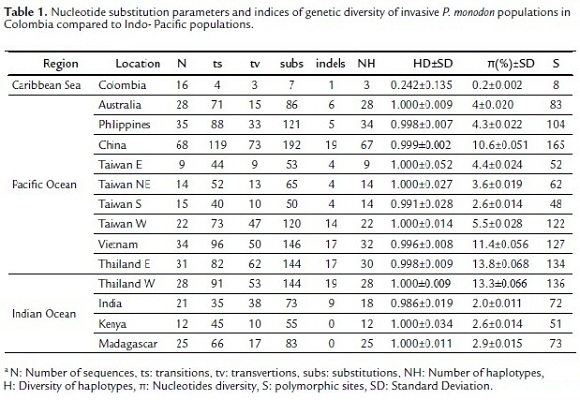
The genetic status of invasive tiger shrimp in Colombia and Indo-Pacific populations was determined by estimating the nucleotide substitution parameters (percentage of nucleotide composition, ts: transitions, tv: transversions, subs: substitutions and indels) and genetic diversity indices (HD: haplotype diversity, π: nucleotide diversity and S: polymorphic sites) with the program Arlequin 3.1.1 (Excoffier et al., 2006).
Possible source of invasive P. monodon in Colombia
The possible source population of the invasive tiger shrimp in Colombia was assessed by determining the genetic relationships between Colombian and Indo-Pacific populations. For this purpose, a Neighbor-Joining tree was constructed in MEGA v5.02 (Tamura et al., 2011). A genetic distance matrix was calculated with pairwise Φst comparisons and an Analysis of Molecular Variance (AMOVA; Excoffier et al., 1992) was performed assuming all samples from each country as independent populations. All these analyzes were performed using the Tamura-Nei model and correction factor Gamma of 0.51 (TN+G; Gamma = 0.51) in the program Arlequin 3.1.1 (Excoffier et al., 2006). The program Modeltest 3.06 (Posada and Crandall, 1998) indicated that the model GRT+G+I was ideal for the control region of the mitochondrial DNA. However, due to the limitations of the software a different yet similar model TN+G was used, a part of Arlequín 3.1.1 (Excoffier et al., 1992), which encapsulates the more complex models that are summarized within TN+G. Finally, a haplotype spatial distribution graphic was made in order to understand the nature of the geographical relationship between the haplotypes present in the Colombian population and those of the Indo-Pacific region. To this end, the frequency matrix obtained in Arlequín was used.
RESULTS
Genetic status of the invasive species P. monodon in Colombia
A total of 358 sequences with a length of 490bp were obtained. The percentage of nucleotide composition of the sequences was similar in all populations but the nucleotide substitution parameters (ts, tv, subs and indels) differed greatly. The Colombian population presented lower substitution values (ts = 4, tv = 3, subs = 7 and indels = 3), whereas the Indo-Pacific populations showed a large variability in the nucleotide substitution of sequences (mean values: ts = 69, tv = 33, subs = 102 and indels = 9). The analysis of genetic diversity recorded 303 haplotypes of the 358 sequences used in the study, the Colombia population with 16 sequences recorded only three haplotypes and their genetic diversity was extremely low (HD = 0.242±0.135, π = 0.2%±0.002 and S = 8) compared to Indo-Pacific populations (mean values: HD = 0.998±0.018, π = 6.2%±0.031 and S = 95) (Table 1).
Source of the species invasive P. monodon in Colombia
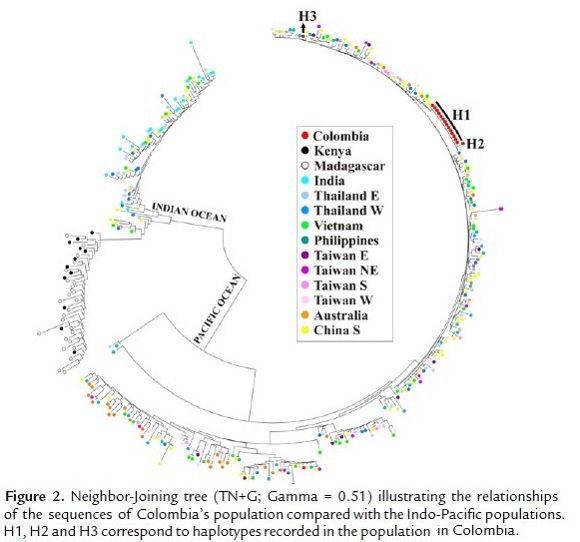
The Neighbor-Joining tree (TN+G; Gamma = 0.51) used to explore the relationships of the Colombian population when compared to Indo-Pacific populations showed that three haplotypes recorded in the 16 sequences are related to haplotypes found in populations from Thailand, Vietnam, Philippines, Taiwan and Australia (Fig. 2).
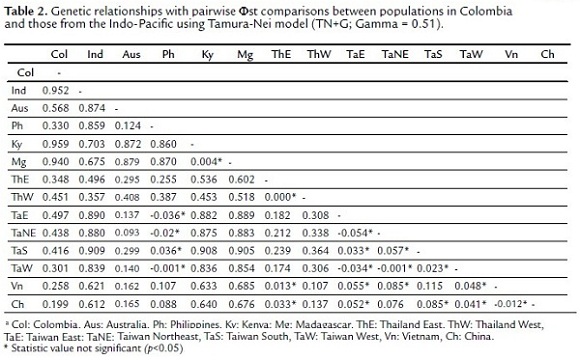
The genetic distance matrix showed a highly significant differentiation between the Colombian population and the Indo-Pacific ones. The lowest values of Φst were registered for China (Col vs Ch = 0.199), Vietnam (Col vs Vn = 0.258), Taiwan (Col vs TaW = 0.301) and Philippines (Col vs Ph = 0.330), whereas there was an increased differentiation with those populations from the Indian Ocean (India, Kenya and Madagascar), even at the species level (Table 2). The AMOVA verified this differentiation among populations (Φct = 0.49, p = 0.0009, Variation = 49.1%) and within populations (Φst = 0.49, p = 0.0000, Variation = 51.0%).
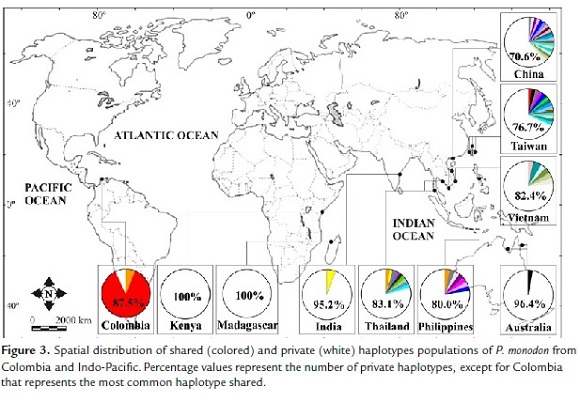
The spatial distribution of haplotypes showed that the most common haplotype in the Colombian population (H1 = 14/16 or 87.5%) was shared with Taiwan, the second one (H2 = 1/16 or 6.25%) was shared with the Philippines and Thailand, and the third one (H3 = 1/16 or 6.25%) was unique (private) to the Colombian groups. However, this doesn't mean that the source of this haplotype (H3) is not the Indo-Pacific. Rather, it means that in order to confirm if this is a real private haplotype, increased sampling of the source populations is needed (Fig. 3).
DISCUSSION
Genetic status of the invasive species P. monodon in Colombia
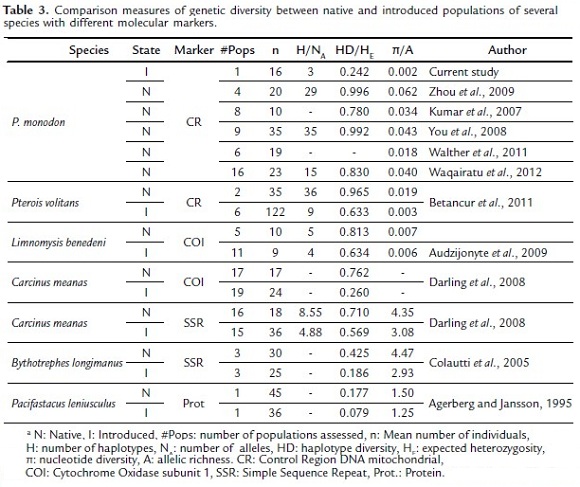
A reduced genetic diversity found in the invasive population of P. monodon in Colombia, compared to native Indo-Pacific populations suggests that the invasion of this shrimp is a result of a founder effect. This means that a few individuals representing a small part of the gene pool of the native population, originated the current invasive population. Even when we compare the data from the invasive population with other studies of native populations (e.g. Kumar et al., 2007; You et al., 2008; Zhou et al., 2009; Walther et al., 2011; Waqairatu et al., 2012), their genetic diversity remains low (Table 3).
Genetic diversity loss associated with a founder effect has been observed in other invading organisms. Several studies in different species using dominant and codominant molecular markers have documented the loss of genetic diversity in introduced organisms compared to their source populations (Table 3). For example, Betancur et al., (2011) using mtDNA to reconstruct the lionfish invasion (Pterois volitans) in the Great Caribbean, found that invasive populations have significantly lower levels of genetic diversity relative to their native counterparts, confirming that their introduction resulted in a strong founder effect. Apparently this is not an uncommon process, because it has always been thought that introduced populations of invasive species should experience loss of genetic diversity relative to native sources because of founder effects and post-introduction demographic bottlenecks (Dlugosch and Parker, 2008).
Source of the invasive Species P. monodon in Colombia
The genetic relationships analysis (pairwise Φst comparisons and Neighbor-Joining tree) showed that the population of tiger shrimp from Colombia is significantly differentiated from populations in the Indian Ocean populations (Kenya, Madagascar and India) and has a lower degree of divergence with those Pacific populations (China, Vietnam, Taiwan and the Philippines, mainly). Although the degree of divergence from the Pacific Ocean populations is high and may be the result of historical processes, this information shows that the origin of invasive tiger shrimp in Colombia is some part of the Pacific Ocean. The spatial distribution of haplotypes allowed establishing their possible origin. The most common haplotype (H1 = 87.5%) is shared with Taiwan and a second (H2 = 6.25%) is shared with the Philippines and Thailand. This shows that the invasive population of P. monodon in Colombia apparently came from these three populations, but mainly from Taiwan. The third haplotype (H3 = 6.25%) was not shared (private haplotype), indicating the likely introduction from another population from sites not included in this study.
However, there is no way to point out the entry process that led to the invasion. They have raised three possible scenarios: 1.The escapement from aquaculture facilities; 2. Migrations from areas where tiger shrimp have previously become established in the wild; and 3.Through ballast water discharge. However, genetic conditions as poor as that found in the population of P. monodon in Colombia may indicate an invasion caused by leakage from farming systems. It is well known that captive populations for commercial purposes are formed by founder effect, that always begin with an artificial bottleneck, where a few individuals are recruited from the natural population (representing only a small part of the gene pool) and are then subjected to selective crosses, causing the loss of genetic diversity due to consanguinity and genetic drift. It is also likely that the individuals of the invading population did not suffer from a single founding effect, but in fact a double effect due to the fact that the shrimp that colonized the Colombian coastal waters originated from cultivation systems, already having initially suffered a first effect.
Benzie (2000) provides evidence of the above when comparing the loss of genetic diversity in wild and cultured populations of the genus Penaeus (including to P. monodon) with several markers such as isoenzymes, RAPD and mtDNA. Even, Sbordoni et al. (1987) recorded decline in observed heterozygosity in seven generations (F1 = 0.102 – F7 = 0.018) on P. japonicus stocks that had been introduced into Italy, reducing hatching rate of 50% to 10% as a result of inbreeding because of the founder effect.
Furthermore, there is evidence that Colombia imported a stock of P. monodon for aquaculture from Brazil in 1987 (Álvarez-León and Gutiérrez-Bonilla, 2007) and the latter made two introductions of this species: the first, from Philippines in the 70's by Empresa de Pesquisa Agropecuária do Estado do Rio Grande do Norte (EMPARN: a State research division dedicated to farming, cattle raising and aquaculture studies) (Madeiros et al., 2006; Leão et al., 2011) and the second, from Taiwan in 1984 by a cultivation enterprise in Valença (Bahia STATE) (Madeiros et al., 2006).
The first report of its increasingly common occurrence in the fisheries of South American coasts appears to be consistent with this introduction period. For example, Fausto-Filho (1987) described the presence of this species in the coastal waters of states from Santos and Maranhão (Brazil), and proposed that their occurrence in the natural environment could be linked to the escape of culture facilities nearby. Likewise, Altuve et al., (2008) considered the possibility that the species escaped to the environment during this initial phase and reached the Brazilian Northeast Atlantic Ocean; later their larvae dispersed northwards with the Guyana current and arrived in the east coasts of Venezuela and Colombia. These authors base their hypothesis on report catches in the French Guiana coast, near the Brazilian border.
However, it can't be ruled out the existence of another processes, like the introduction through ballast water, pathways or migration from other areas of establishment following ocean currents. According Tavares and Braga (2004), P. monodon and other five species of Indo-Pacific native crustaceans, reached the eastern Mediterranean through the Suez Canal, and subsequently were transported in ballast waters to the Western Atlantic. Similarly, Campos and Turkay (1989) suggested this path for the crab Charybdis hellerii, a native species from the Indo-Pacific and first recorded for the Western Atlantic in Bahía Portete (La Guajira, Colombia).
The establishment risk of invasive species P. monodon in Colombian Caribbean
According to Crooks and Rilov (2009) during the invasion process, the establishment stands at the interface between the initial introduction of invaders and their integration into the ecological community. Although the edges of this transitional phase tend to blur, invader establishment is generally related to the survival of initially transported individuals to form reproducing and expanding populations, influenced both by the characteristics of the invader and the receiving ecosystem.
Penaeus monodon, is a species with a wide range of tolerance to changes in environmental conditions. It can live in freshwaters and even in areas where salinities exceed 35 ‰. The species withstands wide ranges in temperature (13°C–33°C) and dissolved oxygen (<3 and >5 mg/L). It also has a high fertility rate (from 500.000 to 750.000 eggs), a fast growth rate and an aggressive predator behavior, feeding mainly on small crabs, shrimp, bivalves, gastropods, fish and polychaetes (Marte, 1980; Su and Liao, 1986; FAO, 2005). Furthermore, it can occupy numerous ecosystems such as lagoons, estuaries, mangroves, the intertidal zone, coral reefs and muddy or sandy bottoms. All these features, combined with their large size (33 cm aprox.) and the lack of predators and challengers, make it a successful organism able to invade new habitats and niches and to establish easily.
Although nothing is known about the impact these shrimp may have on native biodiversity and ecosystem integrity, in recent years it has become more common to find reports of their presence along the coasts of Brazil, Venezuela, Colombia, Puerto Rico and the United States. Moreover, there is evidence to believe that the species is reproducing and completing all its life history stages in Caribbean waters; for example, several authors (Santos and Coelho, 2002; Aguado and Sayegh, 2007; Altuve et al., 2008) have reported catches of adult individuals in marine environments and young ones in estuarine waters. Even, Aguado and Sayegh (2007) mention the capture of ovigerous females in the Orinoco's delta. These findings show the high probability that the species is established or is being established on the South American coasts, representing a major threat to native biodiversity, since the presence of early life stages in lagoon or estuarine systems could threaten and displace native species (e.g. Xiphopenaeus kroyeri and Farfantepenaeus spp.) as a result of competition for space and food. In addition, large sizes achieved in the adult stage and their generalist habit, may allow easily prey on any native species, including other shrimps (Medellín et al., 2011). The above results suggest that P. monodon could be altering the dynamics of populations of native species, changing the structure and function of ecosystems and causing economic significant losses in small-scale fisheries.
On the other hand, poor genetic diversity observed in the population of P. monodon in Colombia could be one favorable aspect in limiting their potential to reproduce, adapt and survive. In this sense, under a constant and selective pressure (e.g. fishing) it could be easier to control and manage their invasive process. However, considering that the invasive process of this species extends beyond the local level, it is necessary to determine through different markers (mtDNA-CR and microsatellite) several important aspects: 1.Whether such poor state of genetic diversity is the same in the population distributed throughout the Caribbean, 2. Whether there is one or several distinct populations and 3. Whether the invasion was produced by a single or by multiple introductions, as the emergence of novel genotypes combinations can promote accelerated rates of evolution and expansion.
It should be considered that the loss of genetic diversity caused by bottlenecks during a founder effect does not always produce negative effects for adaptation and survival of populations. According to Lee (2002) and Barton and Turelli (2004) bottlenecks may create opportunities for new allelic combinations, converting genetic variation due to epistatic or dominance interactions into additive genetic variation. Therefore, increased additive genetic variance during invasion would increase heritable phenotypic variation, providing fuel for evolutionary adaptation in novel environments (Lee, 2002). Also, another important factor to consider is the type of introduction through which the invasion took place, because a single introduction of a large number of individuals or multiple introductions can reduce bottleneck effects as they provide gene flow between source and destinations. Furthermore, the crossing of populations of distinct origin is characterized by an increase in genetic diversity in the invading group (Kolbe et al., 2004). These processes give rise to new genotypes that are possibly more adapted to the new environment and that also promote accelerated rates of evolution and ranges of expansion.
Finally, it is necessary to supplement this information with other ecological studies documenting aspects such as their life history, feeding habits, current population status (abundance and colonization), and fisheries aspects, among others, throughout the area of invasion. Likewise, we discuss the need to create a regional network to further characterize genetically established populations in the Great Caribbean to infer processes of colonization and establish management measures.
CONCLUSIONS
Penaeus monodon is a native species of the Indo-Pacific. It was imported for cultivation in several countries around the world and currently is found on natural environment in the Atlantic Ocean (eg. Mexico, USA, Puerto Rico, Brazil, Guyana, Venezuela), including the Colombian Caribbean Sea. Despite the risk that could be represented, no studies have been conducted to evaluate basic aspect their invasive process in order to understand source and genetic status. This study reveled that Colombia's population had low genetic diversity compared to its native counterpart of Indo-Pacific, suggesting that the introduction was possibly due to a founder effect, related to cultivation attempts of the species, where the specimens apparently come from Taiwan and the Philippines and they were imported by Brazil for cultivation. We discuss the need to create a regional network to characterize the established populations in the Great Caribbean, with the purpose of inferring colonization processes and the establishment of management measures.
ACKNOWLEDGEMENTS
We are indebted to the fishing communities from Pueblo Viejo and Ciénaga. Thanks also to Grupo de Investigación en Biodiversidad y Ecología Aplicada (GIBEA) for their support for laboratory analyses.
REFERENCES
Agerberg A, Jansson H. Allozymic comparisons between 3 subspecies of the fresh-water crayfish Pacifastacus leniusculus (Dana), and between populations introduced to Sweden. Hereditas. 1995;122(1):33-39. Doi: 10.1111/j.1601-5223.1995.00033.x. [ Links ]
Aguado GN, Sayegh J. Presencia del camarón tigre gigante Penaeus monodon (Crustacea, Penaeidae) en la costa del Estado Anzoátegui, Venezuela. Bol Inst Oceanogr Venez. 2007;46(2):107-111. [ Links ]
Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Blackwell Publishing, Oxford, UK; 2007. p. 642. [ Links ]
Álvarez-León R. Antecedentes y posibilidades para el desarrollo de la acuicultura en Colombia. Rev Lat Amer Acuic. 1982;13:9-19. [ Links ]
Álvarez-León R, Gutiérrez-Bonilla F. Situación de los invertebrados acuáticos introducidos y transplantados en Colombia: Antecedentes efectos y perspectivas. Rev Acad Colomb Cienc. 2007;31(121):557-574. [ Links ]
Altuve DE, Marcan LA, Alió JJ, Blanco-Rambla JP. Presencia del camarón tigre Penaeus monodon (Fabricius, 1798) en la costa del delta del río Orinoco y golfo de Paria, Venezuela. Memoria. 2008;68(169):83-92. [ Links ]
Audzijonyte A, Wittmann KJ, Ovcarenko I, Väinölä R. Invasion phylogeography of the Ponto-Caspian crustacean Limnomysis benedeni dispersing across Europe. Divers Distrib. 2009;15(2):346-355. Doi: 10.1111/j.1472-4642.2008.00541.x. [ Links ]
Avise JC. Phylogeography: the History and Formation of Species. Harvard University Press, Cambridge, MA; 2000. p. 427. [ Links ]
Barton NH, Turelli M. Effects of genetic drift on variance components under a general model of epistasis. Evolution. 2004;58(10):2111-2132. Doi: 10.1163/156854080X00139. Article Stable URL: http://www.jstor.org/stable/3449464. [ Links ]
Benzie JAH. Population genetic structure in penaeid prawns. Aquacult Res. 2000;31(1):95-119. Doi: 10.1046/j.1365-2109.2000.00412.x. [ Links ]
Betancur R, Hines A, Acero A, Ortí G, Wilbur AE, Freshwater DW. Reconstructing the lionfish invasion: insights into Greater Caribbean biogeography. J Biogeography. 2011;38(7):1281-1293. Doi: 10.1111/j.1365-2699.2011.02496.x. [ Links ]
Briggs M, Funge-Smith S, Subasinghe RP, PHILLIPS M. Introducciones y movimiento de dos especies de camarones peneidos en Asia y el Pacífico. FAO Documento Técnico de Pesca. Roma; 2005. p. 86. [ Links ]
Campos NH, Turkay M. On a record of Charybdis hellerii from the Caribbean coast of Colombia. Senckenb Marit. 1989;20(3/4):19-123. [ Links ]
Chu KH, Li CP, Tam YK, Lavery S. Application of mitochondrial control region in population genetic studies of the shrimp Penaeus. Mol Ecol Notes. 2003;3(1):120-122. [ Links ]
Coelho PA, Santos MCF, Ramos-Porto M. Ocorrência de Penaeus monodon Fabricius, 1798, no litoral dos estados de Pernambuco e Alagoas (Crustacea, Decapoda, Penaeidae). Bol Técn Cient. CEPENE. 2001;9(1):149-153. [ Links ]
Colautti RI, Manca M, Viljanen M, Ketelaars HA, Bürgi H, Macisaac HJ, et al. Invasion genetics of the Eurasian spiny waterflea: evidence for bottlenecks and gene flow using microsatellites. Mol Ecol. 2005;14(7):1869-1879. [ Links ]
Crooks JA, Rilov G. The Establishment of Invasive Species. In: Rilov G, Crooks JA, editors. Biological Invasions in Marine Ecosystems: Ecological, Management, and Geographic Perspectives. Springer, Berlin, Germany; 2009. p. 173-175. [ Links ]
Darling JA, Bagley MJ, Roman J, Tepolt CK, Geller JB. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus. Mol Ecol. 2008;17(23):4992-5007. Doi: 10.1111/j.1365-294X.2008.03978.x. [ Links ]
Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol. 2008;17(1):431-449. [ Links ]
Excoffier L, Laval G, Schneider S. Arlequin (version 3.1.1): An integrated software package for population genetics data analyses. Evol Bioinform Online. 2006;1:47-50. [ Links ]
Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479-491. [ Links ]
Fausto-Filho J. Registro da captura de Penaeus monodon Fabricius no litoral do estado do Maranhão, Brasil (Crustacea: Penaeidae). Arq Cienc Mar. 1987;26(1):81-82. [ Links ]
FAO. Cultured Aquatic Species Information Programme. Penaeus monodon. Fisheries and Aquaculture Department [online]. Rome. Updated 29 July 2005. [Cited 27 August 2014]. Avaliable at: http://www.fao.org/fishery/culturedspecies/Penaeus_monodon/en. [ Links ]
Frankham R, Ballou JR, Briscoe DA. Introduction to Conservation Genetics. Cambridge University Press, Cambridge, England; 2002. p. 617. [ Links ]
Garcia-Ramos G, Rodriguez D. Evolutionary speed of species invasions. Evolution. 2002;46(4):661-668. Doi: http://dx.doi.org/10.1554/0014-3820(2002)056[0661:ESOSI]2.0.CO;2. [ Links ]
Gómez-Lemos L, Campos N. Presencia de Penaeus monodon Fabricius, 1798 (Crustacea: Decapoda: Penaeidae) en aguas de La Guajira Colombiana. Bol Invest Mar Cost. 2008;37(2):221-225. [ Links ]
Hall T. BioEdit version 5.0.6. North Carolina State University, Department of Microbiology, Unites Estates. 2001. [ Links ]
Holland BS. Genetics of marine bioinvasions. Hydrobiologia 2000;420(1):63-71. Doi: 10.1023/A:1003929519809. [ Links ]
Hulata G. Genetic manipulations in aquaculture: a review of stock improvement by classical and modern technologies. Genetica. 2001;111(1-3):155-73. Doi: 10.1023/A:1013776931796. [ Links ]
Knott DM, Fuller PL, Benson AJ, Neilson ME. Penaeus monodon. USGS Nonindigenous Aquatic Species Database, Gainesville, FL. Revision Date: 6/5/2012. Available at: http://nas.er.usgs.gov/queries/factsheet.aspx?SpeciesID=1209. [ Links ]
Kolbe JJ, Glor RE, Schettino LRG, Lara AC, Larson A, Losos JB. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431(7005):177-181. Doi: 10.1038/nature02807 [ Links ]
Kumar N, Lakra WS, Majumdar KC, Goswami M, Ravinder K. Genetic diversity in the Indian population of Penaeus monodon (Fabricius, 1798) as revealed by mtDNA sequence analysis. Aquacult Res. 2007;38(8):862-869. Doi: 10.1111/j.1365-2109.2007.01740.x. [ Links ]
Leão TCC, Almeida WR, Dechoum M, Ziller SR. Espécies Exóticas Invasoras no Nordeste do Brasil: Contextualização, Manejo e Políticas Públicas. Centro de Pesquisas Ambientais do Nordeste e Instituto Hórus de Desenvolvimento e Conservação Ambiental. Recife, PE. 2011; p. 33. [ Links ]
Lee CE. Evolutionary genetics of invasive species. Trends Ecol Evolut. 2002;17(8):386-391. Doi: 10.1016/S0169-5347(02)02554-5. [ Links ]
Lockwood JL, Cassey P, Blackburn T. The role of propagule pressure in explaining species invasions. Trends Ecol Evolut. 2005;20(5):223-229. Doi: http://dx.doi.org/10.1016/j.tree.2005.02.004. [ Links ]
Madeiros GF, Medeiros LS, Henriques DMF, Lima MT, Genilza C, De Souza VB, Lopes RM. Current distribution of the exotic copepod Pseudodiaptomus trihamatus wright, 1937 along the Northeastern Coast of Brazil. Braz J Oceanogr. 2006;54(4):241-245. Doi: http://dx.doi.org/10.1590/S1679-87592006000300008. [ Links ]
Marte CL. The food and feeding habit of Penaeus monodon Fabricius collected from Makato River, Aklan, Philippines (Decapoda Natantia). Crustaceana. 1980;38(3):225-236. Doi: 10.1111/j.0014-3820.2004.tb01591.x. Article Stable URL:http://www.jstor.org/stable/20103508. [ Links ]
Medellín J, Gómez-Lemos L, Gracia A. Fichas de especies Introducidas. 2011; p. 70-72. In: Gracia A, Medellín-Mora J, Gil-Agudelo DL, Puentes V. Guía de las especies introducidas marinas y costeras de Colombia. INVEMAR, Serie de Publicaciones Especiales No. 23. Ministerio de Ambiente y Desarrollo Sostenible. Bogotá, Colombia. 2011; p 136. [ Links ]
Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817-818. doi:10.1093/bioinformatics/14.9.817. [ Links ]
Ramos G. ¿Será este el nuevo "extramarestre"? Otra especie exótica en nuestras aguas. Fuete y Verguilla. 2012;6(2):1-12. [ Links ]
Santos MCF, Coelho PA. Espécies exóticas de camarões peneídeos (Penaeus monodon Fabricius, 1798 e Litopenaeus vannamei Boone, 1931) nos ambientes estuarino e marinho do nordeste do Brasil. Bol Técn Cient CEPNOR. 2002;10(1):209-222. [ Links ]
Sbordoni V, La Rosa G, Mattoccia M, Cobolli-Sbordoni M, De Matthaeis E. Genetic changes in seven generations of hatchery stocks of the kuruma prawn, Penaeus japonicus (Crustacea, Decapoda). In: Selection, hybridization, and genetic engineering in aquaculture. K. Tiews, editor. Heennemann-Berlag, Berling; 1987. p. 143-155. [ Links ]
SIBM–Invemar. Sistema de Información sobre Biodiversidad Marina de Colombia. Base de datos en línea. 2008. URL: http://www.invemar.org.co/siam/sibm/index.htm (Consulted: 12/11/2008). [ Links ]
Silva KCA, Ramos-PortO M, Cintra IHA. Registro de Penaeus monodon Fabricius, 1798, na plataforma continental do estado do Amapá (Crustacea, Decapoda, Penaeidae). Bol Técn Cient CEPNOR. 2002;2(1):75-80. [ Links ]
Stepien CA, Taylor CD, Dabrowska KA. Genetic variability and phylogeographical patterns of a nonindigenous species invasion: a comparison of exotic vs. native zebra and quagga mussel populations. J Evol Biol. 2002;15(2):314-328. Doi:10.1046/j.1420-9101.2002.00385.x. [ Links ]
Stepien CA, Brown JE, Neilson ME, Tumeo MA. Genetic diversity of invasive species in the Great Lakes versus their Eurasian source populations: Insights for risk analysis. Risk Anal. 2005;25(4):1043-1060. [ Links ]
Su MS, Liao IC. Distribution and feeding ecology of Penaeus monodon along the coast of Tungkang, Taiwan. In: Maclean JL, Dizon LB, Hosillos LV, editors. The First Asian Fisheries Forum: proceedings; 1986 May 26-31; Manila, Philippines. Manila: Asian Fish. Soc. 1986; p. 207-210. [ Links ]
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28(10):2731-2739. Doi: 10.1093/molbev/msr121. [ Links ]
Tavares M, Braga J. Introdução de crustáceos decápodes exóticos no Brasil: Uma roleta ecológica. In: Salles J, Corrêa RS, editors. Água de lastro e bioinvasão. InterCiência, Rio de Janeiro; 2004. p. 59-76. [ Links ]
Tsutsui ND, Case TJ. Population genetics and colony structure of the Argentine ant (Linepithema humile) in its native and introduced ranges. Evolution. 2001;55(5):976-985. Doi: 10.1554/0014-3820(2001)055[0976:PGACSO]2.0.CO;2. [ Links ]
Walther E, Schöfl G, Mrotzek G, Haryanti, Sugama K, Saluz HP. Paralogous mitochondrial control region in the giant tiger shrimp, Penaeus monodon (F.) affects population genetics inference: A cautionary tale. Mol Phylogenet Evol. 2011;58(2):404-408. Doi: 10.1016/j.ympev.2010.11.028. [ Links ]
Waqairatu SS, Dierens L, Cowley JA, Dixon TJ, Johnson KN, Barnes, et al. Genetic analysis of Black Tiger shrimp (Penaeus monodon) across its natural distribution range reveals more recent colonization of Fiji and other South Pacific islands. Ecol Evol. 2012;2(8):2057-71. Doi: 10.1002/ece3.316. [ Links ]
Williamson M. Biological Invasions. Chapman and Hall. New York; 1996. p. 229-257. [ Links ]
You EM, Chiu TS, Liu KF, Tassanakajon A, Klinbunga S, Triwitayakorn K, et al. Microsatellite and mitochondrial haplotype diversity reveals population differentiation in the tiger shrimp (Penaeus monodon) in the Indo-Pacific region. Anim Genet. 2008;39(3):267-277. Doi: 10.1111/j.1365-2052.2008.01724.x. [ Links ]
Zhou FL, Jiang SG, Jiang YJ, Huang JH, Ma ZM. Population genetic structure of the tiger prawn (Penaeus monodon) in the coastal waters of South China, based on mitochondrial DNA control region sequences. J Appl Ichthyol. 2009;2(4):411-416. Doi: 10.1111/j.1439-0426.2009.01228.x. [ Links ]