Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Acta Biológica Colombiana
Print version ISSN 0120-548X
Acta biol.Colomb. vol.20 no.1 Bogotá Jan./Apr. 2015
https://doi.org/10.15446/abc.v20n1.40993
Nota breve
MOLECULAR VARIABILITY OF THREE GENES OF POTATO VEIN YELLOW VIRUS INFECTING Solanum tuberosum, USING SINGLE STRAND CONFORMATIONAL POLYMORPHISM
Variabilidad molecular de tres genes del virus del amarillamiento de las venas de la hoja de la papa que infecta Solanum tuberosum, mediante la técnica de polimorfismo conformacional de cadena sencilla.
Karen Andrea CUBILLOS-ABELLO1, María Mónica GUZMÁN BARNEY1.
1 Laboratorio de Virus Vegetales. Instituto de Biotecnología. Universidad Nacional de Colombia. Bogotá, Colombia.
For correspondence. mmguzmanb@unal.edu.co
Received 27th November 2013, Returned for revision 13th May 2014, accepted 13th October 2014.
Citation / Citar este artículo como: Cubillos-Abello KA, Guzmán-Barney MM. Molecular variability of three genes of Potato vein yellow virus infecting Solanum tuberosum using single strand conformational polymorphism. Acta biol. Colomb. 2015; 20(1):233-237. doi: http://dx.doi.org/10.15446/abc.v20n1.40993.
ABSTRACT
Potato yellow vein virus (PYVV), a virus with tripartite RNA (ss+) genome is classified as member of the genus Crinivirus within the family Closteoviridae. PYVV is the causal agent of potato yellow vein disease (PYVD) with yield loss between 25 %–50 % in field. Single strand conformational polymorphism (SSCP) has been reported to estimate variability in different viruses' species. In this study, the molecular variability of PYVV analyzed by SSCP patterns of three genes: major capsid protein (CP), minor capsid protein (CPm), and heat shock protein (Hsp70) obtained from 60 virus isolates from potato plants expressing PYVD. Leaves collected in Nariño, Colombia from 30 Solanum tuberosum Phureja Group (PhG) and 30 from the Andigena Group (AG). Genes amplified by RT-PCR and the purified PCR products used for SSCP. Three SSCP patterns detected for the CP gene, 12 for CPm and 12 for Hsp70. The pattern C was the most frequent for the Hsp70 gene and the pattern IV (33.3 %) for CPm gene in the Andigena Group. The pattern 1 for CP gene was present in 93.3 % of both host groups, indicating low number of variants compared to the CPm and Hsp70 genes. This is the first attempt to estimate intra e inter PYVV variability by the use of a simple molecular method considering three genes in a large group of potato samples affected by PYVD. SSCP technique was useful to evaluate the viral variability.
Keywords: Closteroviridae, potato, variability, viral proteins.
RESUMEN
Potato yellow vein virus o virus del amarillamiento de venas de la hoja de la papa (PYVV) es un virus RNA tripartito (ss+) de la familia Closteroviridae género Crinivirus que causa la enfermedad de amarillamiento de nervaduras de la hoja de papa (PYVD) reduciendo la productividad, entre el 25 % y 50 %. La técnica de polimorfismo conformacional de cadena sencilla (SSCP) ha sido usada para estimar la variabilidad en diferentes especies de virus. En el presente trabajo se analizó la variabilidad molecular de 60 aislados de PYVV a partir de la comparación de tres genes: el gen de la proteína mayor de la cápside (CP), proteína menor de la cápside (CPm) y la proteína de choque térmico (Hsp70). Los aislados se obtuvieron de dos grupos de Solanum tuberosum: 30 del Grupo Phureja (GPh) y 30 del Grupo Andígena (GA), provenientes del departamento de Nariño, Colombia. Los genes se amplificaron por RT-PCR y los productos purificados se emplearon para SSCP en geles de poliacrilamida. Se detectaron tres perfiles de SSCP para el gen CP, 12 para el CPm y 12 para el Hsp70. Para el GA el perfil más frecuente del gen Hsp70 fue el perfil C (66,6 %) y para el gen CPm fue el IV (33,3 %). Para el gen CP, el patrón uno se encontró en el 93,3 % de los aislados, indicando menor variabilidad. Esta es la primera estimación de variabilidad de PYVV en diferentes genes y a través de una técnica molecular simple.
Palabras clave: Closteroviridae, papa, proteínas virales, SSCP, variabilidad.
RNA viruses characterized by high genetic variability, which is attributed mainly to the absence of RNA dependent RNA polymerase (RdRp) repair activity (Domingo and Holland, 1997). The viruses arises their genetic variation and adaptation through mutation and recombination mechanisms. Variability among members of the family Closteroviridae has been investigated (Rubio et al., 2001; Lozano et al., 2009; Rodríguez et al., 2009). The heat shock protein (Hsp70), minor capsid protein (CPm), and coat major protein (CP) genes are the "hallmark" of this family and have been the target for studies of variability and evolution in all family members (Rubio et al., 2001; Martelli et al., 2002; Essakhi et al., 2006; Lozano et al., 2009). Products of these three genes are structural participating in virion stability, cell-to-cell movement and vector transmission (Napuli et al., 2000; Dolja et al., 2006; Alzhanova et al., 2007).
Potato yellow vein virus (PYVV) is a re-emergent and quarantine virus (EPPO. Data sheet on quarentine pest) classified as a member of the Crinivirus genus within the family Closteroviridae (Salazar et al., 2000). PYVV has a tripartite single stranded RNA genome with positive polarity and is limited to the plant phloem (Salazar et al., 2000; Livieratos et al., 2004). It is the causal agent of potato vein yellow disease (PYVD) (Salazar et al., 2000), and affects different potato varieties in Andean countries, such as Colombia, Peru, Venezuela, and Ecuador (Salazar et al., 2000) causing losses of 25- 50 % in yield (Salazar, 1998; Zapata et al., 2004; Guzmán-Barney et al., 2012; Guzmán-Barney et al., 2013).
The PYVV variability studies have been limited to the coat major protein indicating low variability by single-strand conformation polymorphism (SSCP) (Offei et al., 2004), by restriction fragment length polymorphism (RFLP) (Guzman et al., 2006) and CP sequences (Offei et al., 2004; Rodríguez et al., 2009; Rodríguez et al., 2010). Currently the variability of the coat protein detected slightly higher by sequence analysis (Chaves-Bedoya et al., 2013). Considering the low variability reported for PYVV based on the CP gene analysis by different technical approaches, it could be hope a major variability in different genes of PYVV genome. To evaluate this hypothesis, molecular variability of PYVV was investigated in 60 isolates from Phureja and Andigena Groups of Solanum tuberosum harvested in southwestern Colombia. The SSCPs patterns was analyzed for three genes (CP, CPm, and Hsp70) to estimate inter and intra variability.
The SSCP method has been used to estimates the molecular variability of different viruses as Citrus tristeza virus. It is based on the conformational changes acquired by mutant DNA strands that are visualized following migration in a polyacrylamide gel under non-denaturing conditions (Rubio et al., 1996). The uneven migration of DNA strands is evidenced by the presence of two bands with different electrophoretic mobilities. If one of the chains has a mutation it migrates differently in relation to the other chain and could be detected as a variant. PCR products digested with enzymes show more than one fragment with different migration patterns.
Sixty PYVV isolates were obtained from potato leaves exhibiting symptoms of PYVD. The leaves were collected from 30 Solanum tuberosum Group Phureja (PhG) (criolla native) and 30 Group Andígena (AG) plants (yearly potato). The double-stranded viral RNA (dsRNA) was extracted from samples previously pulverized in liquid nitrogen, using phenol-chloroform (1:1) and purification in Sephadex G50 columns (Guzmán et al., 2006).
Specific CP, CPm, and Hsp70 primers were designed based on the alignment of sequences reported in GenBank (GenBank: AJ557128.1, AJ557129.1, AJ508757.2; Livieratos et al., 2004; Rozen et al., 2000) using the PRIMER3 software (Rozen and Skaletsky, 2000). For the CP gene, the forward and reverse primers were 5´ CTCGAGGATCCTCATGGAAATCCGATC 3´ and 5' CTACTCAATAGAT CCTGCTA 3´, respectively. For the CPmgene, the primers were 5´ ATGGATAAGTCTGTTTTAGATG 3´ and 5´ TCAAAAGTTTTGATTCACATTC 3´ and for Hsp70 were 5´ TGCCCTCTATCTTCAATACCAG 3´ and 5´ CACTTCAAAAATTATCCTACAAAGTGA 3´. To sequence the CPm gene, an internal primer 5´ TCTCTCCAGATCAGGCCAAT 3´ was used. The size of the expected amplified fragments was 768, 2,024 and 1,664 base pairs for the CP, CPm and Hsp70 genes, respectively.
RNA was amplified using two-step RT-PCR. RT reaction consisted of 8 μL RNA, 20 U/μL MMLV reverse transcripatase, 4 U/μL 1X Buffer MMLV, 1 mM dNTPs, and 0.24 μM reverse primer in a final volume of 25 μL. This reaction was performed at 42 °C for 1 h. For the PCR, 4 μL of the RT reaction, 3.75 U/μL Taq polymerase Accuzyme, 1.875 U/μL Biolase, 1X Accuzyme Buffer, 1 mM MgCl2, 0.4 mM dNTPs, and 0.4 μM of each primer was used. The final volume of the reaction was 25 μL. Amplification conditions were 1 cycle at 94 °C for 3 min, 35 cycles at 94 °C for 1 min, 55 °C for 1 min, and 68 °C for 1 min for the Hsp70 and CP genes and 1.30 min for the CPm gene. Finally, an extension at 68 °C for 10 min was performed. The PCR products were purified using the Wizard® SV Gel kit and the PCR Clean-Up System (Promega) and quantified using the QubitTM dsDNA HS kit (Invitrogen). The amplified CPm (2.024 nt) and Hsp70 (1.664 nt) gene products were digested using restriction enzymes to produce shorter fragments in order to obtain higher resolution of migration products using the SSCP method (Rubio et al., 1996). The Hsp70 gene was digested using EcoRI to produce two fragments of 924 and 740 nucleotides. The CPm gene was digested with BstYI to produce four fragments of 636, 528, 622, and 239 nucleotides. The CP gene was not digested. The digestion was performed for 4 h at 37 °C.
For each gene, 3 μL (approximately 6 ng/μL) of the PCR product was collected and mixed with 7 μL of denaturing loading buffer (95 % formamide, 20 mM EDTA, and 0.05 % xylencyanol and bromophenol blue). The samples were denatured by boiling (92 °C for 10 min followed by ten min on ice) and separated by electrophoresis in 8 % polyacrylamide acrylamide-bisacrylamide (29:1) gels under non-denaturing conditions. The CP gene was electrophoresed for four hours, the Hsp70 gene for 4.5 hours, and the CPm gene for three hours, according to the size of the PCR products. The gels were run at a constant 200v at 4 °C (Chare and Holmes, 2004; Guzman et al., 2006) and stained using silver nitrate following the Beidler method (Beidler et al., 1982). The SSCP patterns specific for each gene were analyzed using Quantiti one 4.1 software (BioRad) according to the number of bands (presence or absence), which indicates variability between different virus isolates present in each plant sample. Profiles were classified according to the gene for each host as follows: for the CP gene Arabic numbers (1, 2, 3, etc.); for the Hsp70 gene capital letters (A,B,C, up to N) and for the CPm gene Roman numerals (I, II, III, up to XI). The λ HindIII molecular weight marker was used as a reference for migration in the polyacrylamide gel.
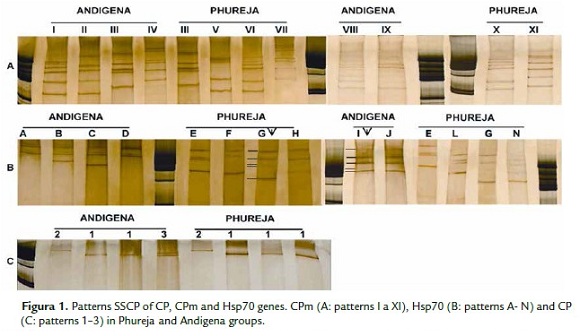
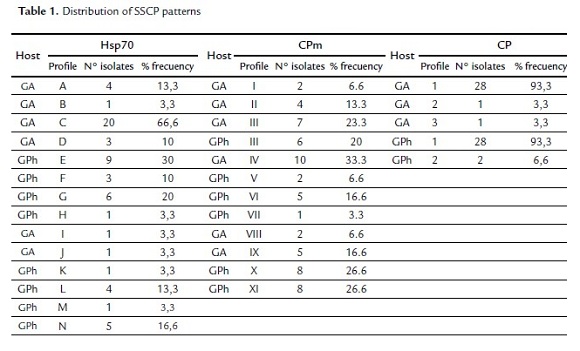
The different SSCP patterns (simple and complex) determined for each gene are illustrated in Fig.1. Six different patterns were obtained for each host in the CPm gene (PhG host: III, V, VI, VII, X, XI and AG host: I, II, III, IV, VIII, IX), only the pattern III was common in both hosts. Eight different patterns were obtained for the PhG host: E,F,G,H,K,L,M,N and 6 different patterns for the AG hosts: A,B,C,D,I, J (Fig. 1B) in the Hsp70 gene. For the CP gene, only three simple patterns were detected, as illustrated Fig. 1C (patterns 1-3). The distribution of each SSCP pattern, related to each gene and host are summarized in the table 1. The pattern most frequent for the Hsp70 gene was C (66.6%), the pattern IV (33.3%) for the CPm gene and the pattern 1 was present in 93.3% of the isolates for the CP gene.
The comparison of SSCP patterns indicated the highest variability in the CPm gene in both hosts, followed by Hsp70 and the CP gene exhibiting the lowest variability.
The SSCP patterns found in this study does not suggest the filtering of a specific variant PYVV into a specific host but rather the random distribution of variants, because different patterns are distributed in the two hosts.
This is the first attempt to evaluate variability of PYVV regarding the migration patterns of three genes biologically important for the virus, due to participate in protection, cell-to-cell movement and vector transmission. The variability data of PYVV using different genetic information is interesting for future improvement programs and biological and evolution information. SSCP is easy to perform and gives a first view of virus variability. Nevertheless, further studies with samples from other Colombian regions will be important to determine the possible presence of more SSCP profiles for each gene, their variability, and their relationship with symptom expression and host specificity.
ACKNOWLEDGEMENTS
We are grateful to Colciencias, project 115-2007, Ministerio de Agricultura y Desarrollo Rural (MARD) 2007S465469 and Dr. Liliana Franco (Universidad Militar Nueva Granada). Patricia Rodríguez cPh.D for her advices about the SSCP technique (Laboratorio de virus de plantas, IBUN-Universidad Nacional de Colombia).
REFERENCES
Alzhanova DV, Prokhnevsky AI, Peremyslov VV, Dolja VV. Virion tails of Beet yellows virus: Coordinated assembly by three structural proteins. Virology. 2007;359(1):220-226. Doi: 10.1016/j.virol.2006.09.007. [ Links ]
Beidler JL, Hilliard PR, Rill RL. Ultrasensitive staining of nucleic acids with silver. Anal Biochem. 1982;126: 374-380. Doi: http://dx.doi.org/10.1016/0003-2697(82)90530-9. [ Links ]
Chaves-Bedoya G, Guzmán-Barney M, Ortiz-Rojas L. Genetic heterogeneity and Evidence Of Putative Darwinian Diversifying Selection In Potato Yellow Vein Virus (PYVV). Rev Agronom Colomb. 2013;31(2):161-168. [ Links ]
Dolja VV, Kreuze JF, Valkonen JP. Comparative and functional genomics of closteroviruses.Virus Res. 2006;117(1):38-51. Doi: http://dx.doi.org/10.1016/j.virusres.2006.02.002. [ Links ]
Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol.1997;51:151-178. Doi: http://dx.doi.org/10.1146/annurev.micro.51.1.151. [ Links ]
Chare ER, Holmes EC. Selection pressures in the capsid genes of plant RNA viruses reflect mode of transmission. J Gen Virol. 2004;85:3149-3157. Doi: http://dx.doi.org/10.1099/vir.0.80134-0. [ Links ]
Essakhi S, Elbeaino T, Digiaro M, Saponari M, Martelli GP.Nucleotide sequence variations in the HSP70 gene of Olive Leaf Yellowing-Associated virus. J Plant. Pathol. 2006;88:285-291. [ Links ]
Guzmán-Barney M, Ruiz E, Arciniegas N, Coutts RH. Occurrence and Variability of Potato yellow vein virus in three Departments of Colombia. J Phytopathol. 2006;154:748-750. Doi: http://dx.doi.org/10.1111/j.1439-0434.2006.01174.x. [ Links ]
Guzmán-Barney M, Franco-Lara L, Rodríguez D, Vargas L, Fierro JE. Yield Losses in Solanum tuberosum Group Phureja Cultivar Criolla Colombia in Plants with Symptoms of PYVV in Field Trials. Am J Pot Res. 2012;89(6):438-447 Doi:10.1007/s12230-012-9265-0. [ Links ]
Guzmán-Barney M, Hernandez A, Franco-Lara L. Tracking Foliar Symptoms Caused by Tuber-Borne Potato Yellow Vein Virus (PYVV) in Solanum Phureja (Juz et Buk) Cultivar "Criolla Colombia". Am J Pot Res. 2013;90(3):284-293. Doi: 10.1007/s12230-013-9303-6. [ Links ]
Livieratos IC, Eliasco E, Muller G, Olsthoorn RC, Salazar LF, Pleij CW, et al., Analysis of the RNA of Potato yellow vein virus: evidence for a tripartite genome and conserved 3'-terminal structures among members of the genus Crinivirus. J Gen Virol. 2004;85:2065-2075. [ Links ]
Lozano G, Grande-Perez A, Navas-Castillo J. Populations of genomic RNAs devoted to the replication or spread of a bipartite plant virus differ in genetic structure. J Virol. 2009;83:12973-12983. Doi: http://dx.doi.org/10.1128/JVI.00950-09. [ Links ]
Internationl Committee on Taxonomy of Viruses. 2012. URL: http://ictvonline.org/taxonomyHistory.asp?taxnode_id=20123509&taxa_name=Potato 2013. [ Links ]
Martelli GP, Agranovsky AA, Bar-Joseph M, Boscia D, Candresse T, Coutts RH et al., The family Closteroviridae revised. Arch Virol. 2002;147:2039-2044. Doi: http://dx.doi.org/10.1007/s007050200048. [ Links ]
Napuli AJ, Falk BW, Dolja VV. Interaction between HSP70 homolog and filamentous virions of the Beet yellows virus. Virology. 2000;274:232-239. Doi: http://dx.doi.org/10.1006/viro.2000.0475. [ Links ]
Offei SK, Arciniegas N, Muller G, Guzman M, Salazar LF, Coutts RH. Molecular variation of Potato yellow vein virus isolates. Arch Virol. 2004;149:821-827. Doi: http://dx.doi.org/10.1007/s00705-003-0250-2. [ Links ]
Rodríguez P, Chaves G, Franco L, Guzmán M. Low molecular variability of Potato yellow vein virus (PYVV) isolates of Solanum phureja and Solanum tuberosum from Colombia. Phytopathology. 2010;100:S176. http://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO.2010.100.6.S172. [ Links ]
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365-386. [ Links ]
Rozen S, Skaletsky, Krawetz S, Misener S (eds). Bioinformatics methods and protocols: methods in molecular biology. Humana press NJ. 2000:365-386. [ Links ]
Rubio L, Ayllon MA, Kong P, Fernandez A, Polek M, Guerri J, et al., Genetic variation of Citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. J Virol. 2001;75:8054-8062. Doi: http://dx.doi.org/10.1128/JVI.75.17.8054-8062.2001. [ Links ]
Rubio L, Ayllon MA, Guerri J, Pappu HR, Niblett CL, Moreno P. Differentiation of Citrus tristeza virus (CTV) isolates by single-strand conformation polymorphism analysis of the coat protein gene. Ann App Biol. 1996;129:479-489. Doi: http://dx.doi.org/10.1111/j.1744-7348.1996.tb05770.x. [ Links ]
Salazar LF, Miller G, Querci M, Zapata JL, Owens RA. Potato yellow vein virus: its host range, distribution in South America and identification as a crinivirus transmitted by Trialeurodes vaporariorum. Ann App Biol. 2000;137:7-19. Doi: http://dx.doi.org/10.1111/j.1744-7348.2000.tb00052.x. [ Links ]
Salazar LF. Technology Development for Control of Yellow Vein Disease in Colombia. in Center I.P. (ed), Perú;1998. p.35. [ Links ]
Zapata JL, Saldarriaga A, Salazar LF. El amarillamiento de venas de la papa. Corpoica. Rionegro, Antioquia. 2004;1-12. [ Links ]