INTRODUCTION
Larvae from benthic marine invertebrates are able to discriminate between different substrata for settlement and metamorphosis, favoring those that provide a higher chance of survival and reproduction (Harrington et al., 2004). In cnidarians, bacterial biofilms established on settling surfaces play a key role on larval attachment and metamorphosis, as it has been shown in the corals Acropora microphthalma (Webster et al., 2004) and Pocillopora damicornis (Tran and Hadfield, 2012). In addition, monospecific biofilms of the bacterium Pseudoalteromonas sp. isolated from crustose coralline algae are able to induce larval settlement and metamorphosis in vitro in the corals Acropora millepora and A. willisiae (Negri et al., 2001), and in the hydroid Hydractinia echinata (Leitz and Wagner, 1993, Seipp et al., 2007). The settlement induction activity of Pseudoalteromonas sp. biofilms has been attributed to the compound tetrabromopyrrole (TBP), and its inductive properties have been demonstrated in larvae from the corals Porites asteroides (Sneed et al., 2014) and Acropora millepora (Tebben et al., 2011). Biofilms, however, potentially produce a vast diversity of molecules that might affect larval substrate selection, most of which are yet to be identified (Hadfield, 2011).
The formation of bacterial biofilms is often modulated by Quorum Sensing (QS), an intercellular communication system based on density-dependent regulation of gene expression (Parsek and Greenberg, 2005). QS regulates a variety of other bacterial activities such as secondary metabolite production, virulence, and motility (Hooi et al., 2004, Williams et al., 2007), and it relies on low molecular weight, diffusible molecules known as Quorum Sensing signaling molecules (QSSMs). The main class of QSSMs in Gram-negative bacteria is the N-acyl-L-homoserine lactones (AHLs), which are a large family of related molecules that differ in length (from 4-18 carbons), substitutions at the acyl side chain (3-oxo, 3-hydroxy, or unsubstituted) and the degree of saturation (Miller and Bassler, 2001). The AHL-dependent QS involves the synthesis of AHLs and their interaction with transcriptional regulators that activate target genes (Williams et al., 2007). Another QSSM in Gram-negative bacteria are the 4-quinolones (Diggle et al., 2006), whereas QSSMs in Gram-positive bacteria are the autoinducing polypeptides (Sturme et al., 2002)2002 and the y-butyrolactones (Takano, 2006).
QSSMs produced by bacterial biofilms affect settlement behaviors in some organisms. For example, migration of spores from the green alga Ulva sp. is facilitated by AHL-producing bacterial biofilms (Tait et al., 2005; Wheeler et al., 2006), whereas larvae settlement in the polychaete Hydroides elegans is positively correlated with the density of AHL-producing bacteria in a biofilm (Huang et al., 2007). In this study, we evaluated the role QSSMs in the settlement and metamorphosis of larvae from the colonial hydroid Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa). This hydroid is found in near-shore waters of the northeastern United States, growing as a surface incrustation on gastropod shells occupied by the hermit crab Pagurus longicarpus (Buss and Yund, 1989). Mature colonies of this dioecious species release their gametes to the water for fertilization, initiating a developmental process that culminate in the formation of a planula larva having an anterior blunt end and a posterior pointed end (Kraus et al., 2014). Following appropriate stimuli, the larvae settle on the gastropod shell by attaching the anterior blunt end to the surface, initiating a 2-3 day metamorphosis process that results in a primary polyp, which sets off the formation of a colony by budding new polyps from extending canals (Berking, 1991). In Hydractinia, the progression from a planula larva to a primary polyp has been divided into 14 morphological stages based on artificial induction of metamorphosis with cesium chloride (Seipp et al., 2007). Stages 1-6 involve a progressive shortening in the larva's anterior-posterior axis until forming a drop-shaped structure (stages 7-8). In stage 9, the drop-shaped larvae adhere to the substratum by the anterior end, and the larva's posterior end rounds (stage 10) to acquire a disc form (stage 11). In stage 12, the metamorphosing structure has a barbell shape and the canal and tentacle buds become visible (stage 13), to finally form a primary polyp in stage 14.
MATERIALS AND METHODS
H. symbiolongicarpus culture and larvae production
We collected colonies of H. symbiolongicarpus growing on gastropod shells occupied by hermit crabs in Woods Hole, Massachusetts, USA. We subsequently explanted colony fragments of 3-5 polyps onto glass microscope slides and maintained them at 20 ± 2° C in 40-liter aquaria with recirculating artificial seawater (Instant Ocean), feeding them to repletion with 2-day old Artemia salina nauplii three times a week. Five male and five female sexually mature colonies were induced to release gametes by exposure to light for 30 min (Ballard, 1942). We collected the fertilized eggs into 35 mm Petri dishes where larvae developed within 36 hr at room temperature. Shells containing the remaining H. symbiolongicarpus colonies were kept in separate aquaria under identical conditions to be used for bacterial isolation.
Bacteria isolation and identification
Two shells with H. symbiolongicarpus were used for bacteria isolation by peeling off part of the colony from the shell and streaking the shells onto Luria Bertani (LB, Gibco) and Marine agar (Gibco) plates. The plates were incubated at 25°C and 37°C for 24 and 48 hours, and the resulting bacterial colonies were systematically purified by serial dilutions in phosphate buffer and re-plated on LB and Marine Agar to be finally preserved in glycerol at -80° C. Each bacterial isolate was characterized morphologically by form, Gram staining, color, texture, margin and elevation, and biochemically by using the standard bacteriological tests, indole production, growth on triple sugar iron agar, growth on lysine iron agar, citrate utilization, nitrates production, motility, carbohydrates utilization, methyl-red Voges-Proskauer reaction and catalase activity (Holt et al., 1994).
The isolates were identified by sequencing the 16S rDNA gene from PCR products generated with the universal primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3'), using the protocols reported by Cárdenas et al., (2012) The resulting sequences of about 1350 bp (GenBank accession numbers KU144692-KU144704) were aligned using the ClustalW software (Thompson et al., 1994), and a preliminary species assignation was carried out based on sequence similarity values of 97-100% against the GenBank and LPSN databases (Stackebrandt and Goebel, 1994; Rosselló-Mora and Amann, 2001), Rosselló-Mora and Amann, 2001. Phylogenetic trees were constructed using the MEGA 5 package (Tamura et al., 2011) with the Neighbor-joining method (Saitou and Nei, 1987) based on distances estimated by the Kimura two-parameter model (Kimura, 1980). The reliability of the tree topology was evaluated by bootstrapping (Felsenstein, 1985) using 1000 replications.
Detection of bacterial QSSMs
The production of QSSMs by the bacterial isolates was evaluated with the cross-streaking technique with four bacterial sensors that respond to AHLs (Steindler and Venturi, 2007): a) E. coli pSB403 and b) E. coli pSB1075, which activate the LuxI/R and LasI/R QS systems, respectively, to produce bioluminescence (Winson et al., 1998), c) Chromobacterium violaceum CV026 which activates the Cvil/R QS system to produce violacein (McClean et al., 1997), and d) Agrobacterium tumefaciens NT1 pZLR4 which uses the TraI/R QS system coupled with of β-Galactosidase enzyme as reporter (Ferrand et al., 2002). The biosensors were kindly provided by Dr. Zulma Suárez (E. coli pSB403 and pSB1075), and by Dr. Katharina Riedel (C. violaceum CV026 and A. tumefaciens NT1 pZLR4 The presence of QSSMs was also evaluated by reverse phase-thin layer chromatography (RP-TLC) on extracts obtained with acidified (0.01 % acetic acid) ethyl acetate following the protocol reported by Ortori (Ortori et al., 2011). For the RP-TLC, C18 plates (aluminum sheets RP-18 F254S , 20 x 20 cm, Merck Chrome Line) were used, with methanol/water (60:40, v/v) as the mobile phase. The plates were covered with a 143 mL of AB medium, seeded with the biosensor A. tumefaciens pZLR4 and 760 μl of X-Gal at 20 mg/ml, and subsequently incubated at 28°C for 24 hours. The strain Pseudomonas putida F117, which is unable to produce AHLs, was used as negative control (Steidle et al., 2002).
Chemotaxis assays with multispecies biofilms
We generated a multispecies biofilm with the three QSSM-producing bacteria S. flexneri, M. oxydans, and K. erythromyxa. The three bacteria strains were cultured independently in a liquid medium with 0.02 % of yeast extract in artificial sea water to an OD600 of 0.2. Five ml of each culture were combined and filtered by vacuum on a 0.45 μm pore nitrocellulose filter of 35 mm of diameter using the method reported by Seipp et al., (2007). In order to enhance its inductive capacity, the biofilm was incubated overnight in artificial sea water at 4° C (Seipp et al., 2007). The activated biofilm was then placed at the center of a 100 mm polystyrene Petri dish containing 17 mL of sterile artificial seawater. One hundred H. symbiolongicarpus larvae of two days post-fertilization were then transferred to the Petri dish in 500 μL of artificial seawater, placing them with a pipette evenly distributed at the circumferential edge of the dish. Larvae movements from the periphery towards the biofilm at the center were registered 1, 3, 6, 9, 12 and 24 hours later according to their location into four concentric regions marked in the Petri dish. As a negative control, 16 mL of sterile media were adsorbed on a nitrocellulose filter by vacuum and the movement of 100 larvae on the Petri dish was evaluated as described before. Significant differences between treatment and control in larvae migration from the periphery to the center of the dish were tested with a Poisson generalized linear model based on three independent assays with 100 larvae each.
Chemotaxis assays with QSSM extracts
QSSM extracts were obtained from 500 mL liquid cultures of S. flexneri, M. oxydans, and K. erythromyxa using acidified ethyl acetate (Ortori et al., 2011) at a final volume of 250 μL. Extracts from each culture were pooled and 12 μL of the pool were combined with 500 μL of 0.5 % low melting point agarose (Life Technologies) in artificial seawater. The agarose plug with the extract was then placed at the center of a 100 mm polystyrene Petri dish with 17 mL of sterile artificial seawater. H. symbiolongicarpus larvae of two days post-fertilization (n = 100) were then placed at the edge of the dish as described before, and larvae counts within the dish's four concentric regions were recorded at 1, 3, 6, 9, 12 and 24 hours after placement. An extract from sterile media was used as negative control, and ethyl acetate extracts from the strains E. coli DH5ce and C. violaceum CV026, which do not produce AHLs, were also used to exclude the effects of non-QSSM on larvae movements. The former is a highly domesticated strain that has lost the biosynthetic machinery to produce AHLs (Surette and Bassler, 1998), whereas the latter is a mini Tn5 mutant strain with a truncated AHL synthase gene (McClean et al., 1997). A Poisson generalized linear model based on three independent assays was used to test differences between treatments and controls.
Induction of larval settlement and metamorphosis with biofilms and QSSM extracts
Multispecies biofilms and QSSM extracts from S. flexneri, M. oxydans, and K. erythromyxa were obtained as detailed above. Biofilms established on 0.45 μm nitrocellulose filters were placed in 35 mm Petri dishes with 5 mL of sterile artificial seawater and 100 H. symbiolongicarpus larvae of two days post-fertilization. For assays with QSSM extracts, 8 mL of sterile artificial seawater were added to each well of a 6-well microtiter plate and supplemented with 12 μL of the QSSM extract, together with 100 larvae of two days post fertilization. In addition, extracts from the AHL-negative strains E. coli DH5α and C. violaceum CV026 were used as controls. Progression of metamorphosis was evaluated at 1, 3, 6, 9, 12 and 24 hours after larvae placement. Larvae were classified into stages 1-6 ("swimming"), stage 7-10 (drop-shaped), stage 11 (attached disc), and stage 14 (primary polyp). As positive control of metamorphosis induction, 100 larvae of 2 days post fertilization were incubated in a solution of 53 mM CsCl in sterile artificial seawater for two hours and subsequently transferred to sterile seawater to allow metamorphosis. Assays were done in triplicate and differences between treatments and controls were evaluated statistically by using the binomial test.
RESULTS
Isolation and identification of bacteria from shells with H. symbiolongicarpus
We obtained 14 bacterial isolates from two shells with H. symbiolongicarpus epibionts. These strains were characterized morphologically and biochemically, and 1,350 bp of the 16S rRNA gene were sequenced for a preliminary taxonomic assignation based on a sequence similarity range of 97100 % against the GenBank and LPSN databases (Table 1). Eleven of the 14 strains were tentatively assigned at the species level, two were assigned to the genus level, and one (CCB-8A) was not even possible to assign at the genus level, as the closer sequence in the databases showed a similarity of 77.6 % (Table 1). A phylogenetic analysis based on the 16S rDNA sequences corroborated the species assignation and revealed that they clustered into three main clades corresponding to the phyla Actinobacteria, Proteobacteria, and Firmicutes, containing the genera Kocuria, Micrococcus, Microbacterium, Shigella, Pantoea, Exiguobacterium, Bacillus, and Staphylococcus (Fig. 1).
Table 1 Morphological, biochemical, and taxonomic characterization of bacterial strains isolated from shells with H. symbiolongicarpus
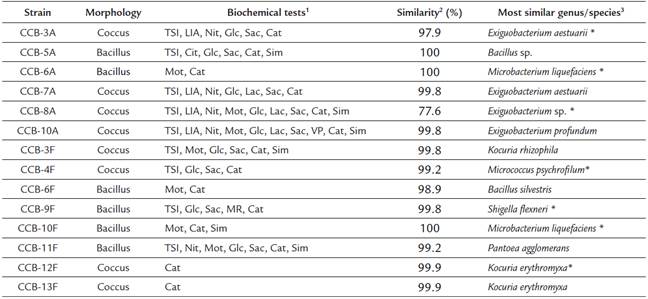
1 Positive results for the biochemical tests, TSI (triple sugar iron), LIA (lysine iron agar), Cit (citrates uptake), Nit (nitrates reduction), Mot (motility test), Glc (glucose degradation), Lac (lactose degradation), Sac (saccharose degradation), MR (methyl red), VP (Vogues-Proskauer), Cat (catalase), and Sim (sulfide indol motility).
2 Percentage of 16S rDNA sequence similarity between the isolated strains and the most similar species in the GenBank and LPSN databases.
3 Strains marked with an asterisk (*) were used for QSSM extraction.
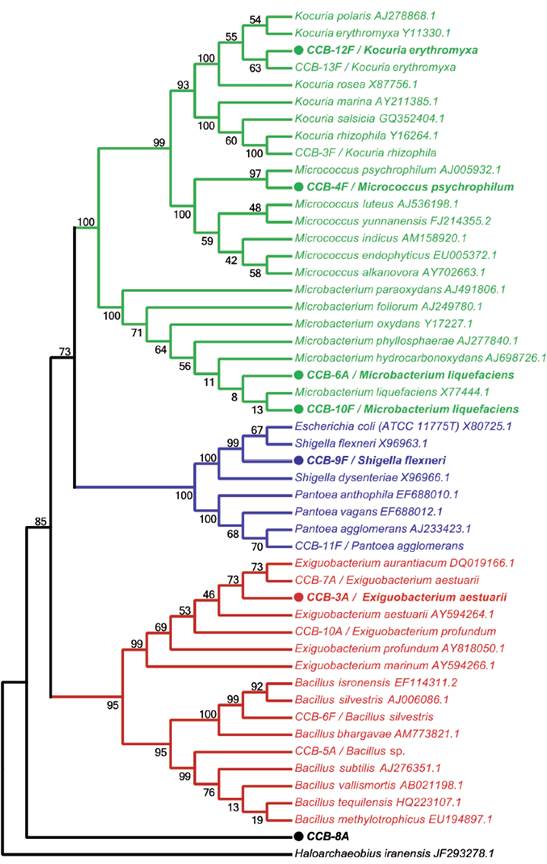
Figure 1 Taxonomic analysis of bacterial strains isolated from shells with H. symbiolongicarpus colonies. The neighbor-joining tree was based on 16S rDNA sequences from all 16 isolated strains together with GenBank sequences from selected species, using Haloarchaeobius iranensis as outgroup. The isolates were named with a unique identifier and a tentatively assigned species name. Branches and labels in red represent species from the phylum Firmicutes, those in blue are species from the phylum Proteobacteria, and in green are represented species from the phylum Actinobacteria. A filled circle preceding the isolate identifier indicates that the strain was tested for the production of QSSMs and numbers on branches indicate bootstrap percentages after of 1000 replicates.
QSSMs were detected in three bacterial isolates
We evaluated seven of the identified species (marked with asterisks in Table 1 and filled circles in Figure 1) for QSSMs production using the cross-streaking assay (Steindler and Venturi, 2007). For this assay, the biosensors employed were the bioluminescent strains E. coli pSB403 and pSB1075, which are activated with exogenous AHLs having short and long acyl side chain, respectively, the violacein-producing C. violaceum CV026 strain, that detects short acyl side chain AHLs, and the β-galactosidase-based sensor Agrobacterium tumefaciens pZLR4, which detects a wide range of AHLs (Steindler and Venturi, 2007). Three of the seven evaluated strains, CCB-9F (Shigella flexneri), CCB-10F (Microbacterium liquefaciens), and CCB-12F (Kocuria erythromyxa), activated two of the four biosensors, E. coli pSB1075 and A. tumefaciens pZLR4 (Fig. 2). The E. coli pSB1075 biosensor detects 3-oxo-C12-AHL, C12-AHL and/or 3-oxo-C10-AHL, whereas the A. tumefaciens pZLR4 biosensor detects C6-AHL, C8-AHL, C10-AHL, C12-AHL and its derivatives with 3-oxo and hydroxyl substitutions (Steindler and Venturi, 2007). The production of QSSMs by S. flexneri, M. liquefaciens, and K. erythromyxa was confirmed by reverse phase-thin layer chromatography (RP-TLC) using acidified ethyl acetate extracts from liquid cultures and the A. tumefaciens pZLR4 biosensor (Figure 3). Based on the spot mobility in the RP-TLC in comparison to the control extract from P. aeruginosa PAO1, it is likely that the detected QSSMs are 3-oxo-C6-AHL in S. flexneri and M. liquefaciens, and 3-oxo-C6-AHL, 3-oxo-C8-AHL, 3-oxo-C10-AHL and 3-oxo-C12-AHL in K. erythromyxa. The lack of detection of QSSMs in the remaining strains is likely due to either their inability to produce or recognize AHLs or to the production of the molecules at low concentrations, undetectable by the system.
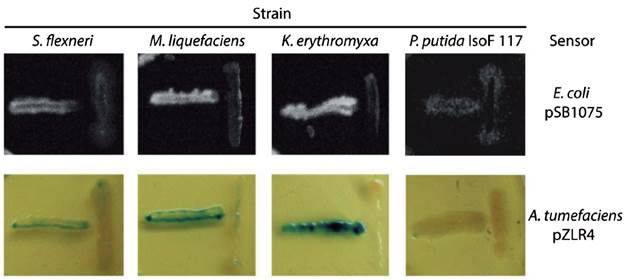
Figure 2 Identification of bacterial strains producing QSSMs using the cross-streaking assay. The biosensors E. coli pSB1075 and A. tumefaciens pZLR4 were confronted with the shell bacterial isolates S. flexneri, M. liquefaciens, K. erythromyxa, and with the negative control P. putida IsoF117. Horizontal streaks correspond to the biosensors and the vertical streaks to the assayed strains. These assays were performed on LB agar plates incubated 16 hr at 37°C for E. coli pSB1075 and at 28°C for A. tumefaciens pZLR4. Bioluminescence was detected using a BIORAD Quantity-One 4.1.0 imager.
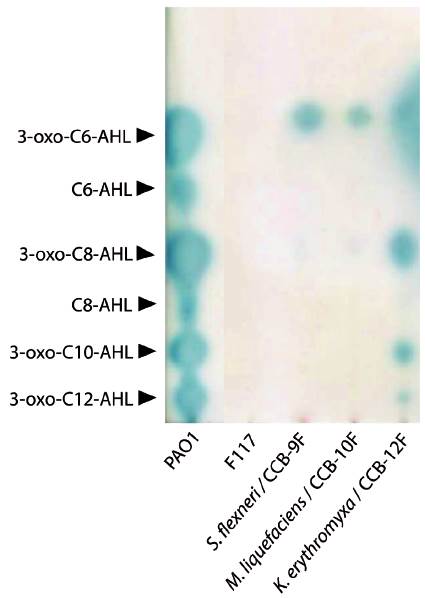
Figure 3 Reverse phase-thin layer chromatography (RP-TLC) of QSSMs extracts from shell-isolated strains. The QSSM extract from the reference strain P. aeruginosa PAO1 was used as mobility pattern whereas the strain P. putida IsoF117 was used as negative control. Samples were spotted on RP-TLC plates, developed with 60:40 methanol: water and overlaid with a suspension of the biosensor A. tumefaciens pZLR4 in soft AB medium with X-Gal. The position and identity of molecules is indicated with arrowheads according to R f values.
Biofilms and QSSM extracts have chemotactic effects on H. symbiolongicarpus larvae
We tested chemotaxis of H. symbiolongicarpus larvae towards a biofilm or a QSSM extract by placing, in triplicate, 100 larvae of two days post-fertilization evenly distributed at the edge of a Petri dish, with the inductor at the center of the dish. Larvae locations within four concentric regions in the dish were recorded after after 1, 3, 6, 9, 12 and 24 hours. Three assays having different chemotactin inductors were employed: a) a multispecies biofilm conformed by S. flexneri, M. liquefaciens, and K. erythromyxa, b) a pool of acidified ethyl acetate extracts from S. flexneri, M. liquefaciens, and K. erythromyxa liquid cultures, and c) acidified acyl acetate extracts from the strain E. coli DH5α and C. violaceum CV026, which are unable to produce QSSMs but may produce other molecules not regulated by QS that could affect larvae migration. As depicted in Figure 4A, there was a clear tendency for H. symbiolongicarpus larvae to migrate towards the multispecies biofilm at the center of the dish, in significant contrast with the control (p < 0.05) where larvae mostly remained at the most peripheral region of the dish (Table 2). In the second assay, the larvae significantly migrated towards the pooled QSSM extracts (p < 0.05) in comparison to the control (Fig. 4B and Table 2). Indeed, at 24 hrs post initiation, in the most peripheral region of the dish with the QSSM extract, there was an average of 7 ± 3.2 % of larvae in comparison with 68.3 ± 5.7 % in the control (Table 2). Finally, in the assay with the extracts from mutant strains unable to produce QSSMs there was no a significant difference between the treatment and the control (Figure 4C and Table 2). These results indicate that biofilms of the bacterial species isolated from the shells have a chemoattractant effect on H. symbiolongicarpus larvae, and that the effect might be attributed to QSSMs in the biofilms and the extracts.
Table 2 Mean number of larvae (± SD) located at each concentric region of the dish at different times post-induction.
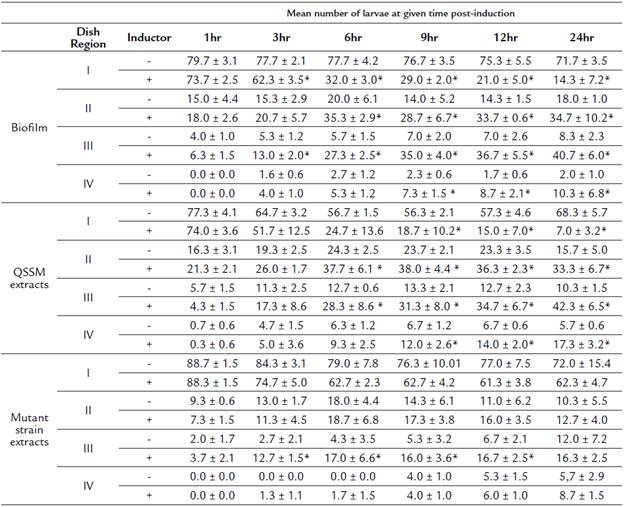
(*) Represent significant differences between treatments using the inductor (biofilm, QSSM extracts or mutant strain extracts) and its respective negative control.
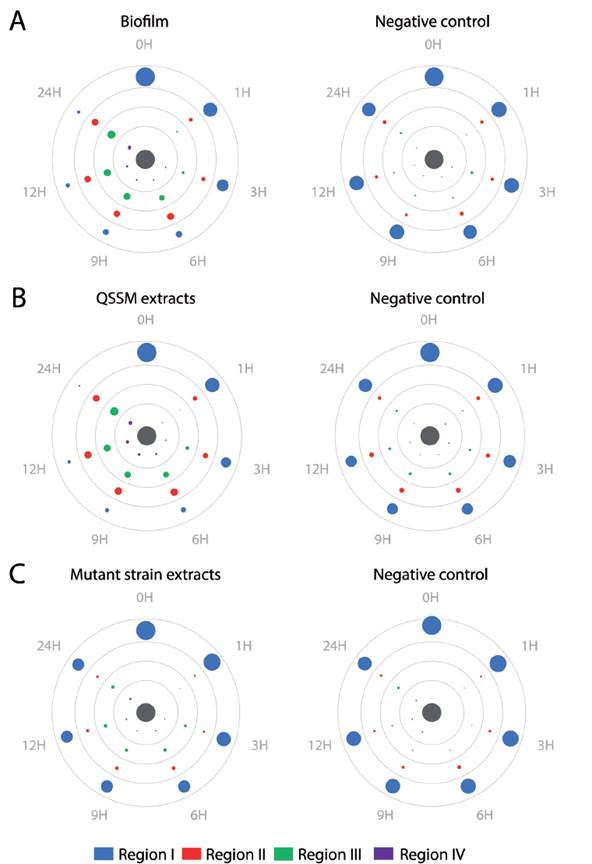
Figure 4 Biofilms and QSSM extracts from shell-isolated bacterial strains induce chemotaxis of H. symbiolongicarpus larvae. One hundred larvae were evenly placed at the periphery of a Petri dish with an chemotactic inductor at the center (grey circle), and their location within four concentric regions were recorded at 1, 3, 6, 9, 12, and 24 hrs. The inductors were (A) a biofilm from the shell-isolated strains S. flexneri, M. liquefaciens, K. erythromyxa, (B) a pooled QSSM extract from the three shell-isolated strains, and (C) a pooled extract from the mutant strains E. coli DH5a and C. violaceum CV026, which are unable to produce QSSMs. Color circles represent the mean number of larva from three independent assays located in one of the four concentric regions, with the circle area proportional to number of larva found at a region in a given time (mean values are shown in Table S1). Negative controls were extracts from non-inoculated media.
QSSM extracts induce larval attachment but not metamorphosis to a primary polyp
We evaluated the multispecies biofilm and the pooled QSSM extracts of S. flexneri, M. liquefaciens, and K. erythromyxa for their capacity to induce metamorphosis in H. symbiolongicarpus larvae. One-hundred larvae were incubated in triplicate with the biofilm or the QSSM extracts, and their metamorphosis progression into stages 1-6 ("swimming"), stage 7-10 (drop-shaped), stage 11 (disc), and stage 14 (primary polyp) were followed for 24 hrs (Fig. 5). The assay with the multispecies biofilm (red bars in Figure 5) showed that at 24 hrs post exposure, an average of 5 ± 3.7 % of larvae were at the swimming stage, 90 ± 2.8 % at the drop-shaped stage, 4 ± 2.8 % at the disc stage, and just 1.3 ± 0.7 % developed into a primary polyp. In contrast, most larvae from the negative control (77.7 ± 4.0 %) remained at the swimming stage, whereas 89 ± 2.9 % of larvae from the positive control (CsCl induction) reached the primary polyp stage.
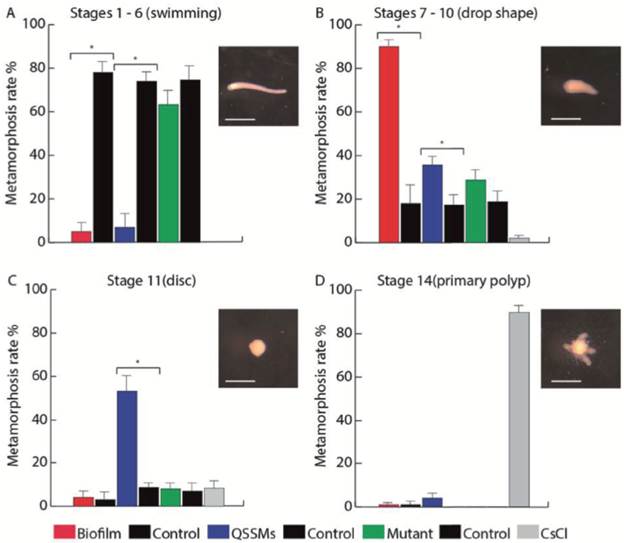
Figure 5 Biofilms and QSSM extracts from shell-isolated bacterial strains induce presettlement and settlement behaviors of H. symbiolongicarpus larvae. Percentage of larvae at (A) the swimming stage, (B) drop shape stage, (C) disc stage, and (D) primary polyp stage, were counted at 24 hrs after the induction with a biofilm and QSSM extract from shell-isolated bacterial strains, and from extracts from the mutant strains E. coli DH5a and C. violaceum CV026. Negative controls were paired with each treatment and extracts from non-inoculated media, and CsCl was used as a positive control of metamorphosis. Bars represent the mean percentage of larvae from three independent experiments with 100 larvae each. Asterisks indicate a significant difference between a treatment and its control (P < 0.01, binomial test). Scale bars in larvae photographs represent 500 μm.
In the assay with the QSSM extracts (blue bars in Figure 5), 5.7 ± 5.2 % of larvae remained at the swimming stage at 24 hr, 37.3 ± 3.3 % at the drop shape stage, 53.3 ± 6.8 % at the disc stage, and 4 ± 2.6% progressed to form a primary polyp. Extracts from mutant strains unable to produce QSSMs (green bars on Figure 6) had no presettlement or settlement inductive activity on larvae, as most of the larvae remained at the swimming stage (63.3 ± 5.3 %), whereas 29.7 ± 4.2 % reached the drop shape stage, showing no significant difference to the negative control. Thus, both the biofilms and the QSSM extracts from the three bacterial species isolated from the shells induced high rates of settlement, although little or no metamorphosis to a primary polyp.
DISCUSION
Bacterial biofilms play a central role promoting the settlement and metamorphosis of many marine invertebrate larvae. Yet, the molecular cues responsible for those behaviors have remained, with some exceptions, elusive. Larvae of the cnidarian H. symbiolongicarpus settle and metamorphose on gastropod shells occupied by hermit crabs, and this system has proved to be an adequate model to investigate the complex relations between bacterial biofilms and the biology of substrate selection (Seipp et al., 2007). In this study, we showed that QSSM extracts from bacterial communities colonizing the gastropod shells influence the settlement of H. symbiolongicarpus larvae. The bacteria species isolated from the shells (S. flexneri, M. liquefaciens and K. erythromyxa) have been previously associated to diverse marine environments (Kim et al., 2005, Crapart et al., 2007), including Caribbean reef-building corals (Cárdenas et al., 2012). These isolates were identified to produce QSSMs, most likely long side chain AHLs as the biosensors employed to detect QSSMs respond specifically to the presence of AHLs, which are the typical QSSMs of Gram-negative bacteria. Although one of the isolated species (M. liquefaciens) is Gram-positive, while another one (K. erythromyxa) is Gram-variable, production of AHLs by Gram-positive bacteria have been reported before (Biswa and Doble, 2013). Yet, the possibility that a chemically different molecule from this Gram-positive bacterium activates AHL-based QS circuitry should not be excluded.
Previous studies evaluating larval settlement cues have suggested that QSSMs might play a role in the phenomenon. For example, in the polychaete Hydroides elegans, QSSMs (AHLs) affect pre-settlement behaviors such as decreasing larval swimming speed or crawling to the bottom of the testing dish, but they do not induce settlement (Huang et al., 2007). Similarly, zoospores of the macroalgae Ulva decrease their swimming speed through a process of cytokinesis (Wheeler et al., 2006) and increase settlement rates in response to AHL-producing biofilms (Tait et al., 2005). In addition, cyprid larvae of the barnacle Balanus improvisus settle as a response of AHL-producing biofilms or synthetic AHLs (Tait and Havenhand, 2013). The present study reports for the first time in a cnidarian the inductive role of QSSM extracts from substrate-specific bacteria on larval presettlement and settlement behaviors. H. symbiolongicarpus larvae consistently migrated towards multispecies biofilms and extracts from the QSSM-producing isolates, whereas they did not show any directional movement towards extracts from mutant bacterial strains unable to produce QSSMs or towards negative controls. Hence, it is likely that QSSMs produced by the bacterial biofilms growing on the shells exert a chemotactic effect on H. symbiolongicarpus larvae.
In Cnidaria, larva settlement involves two independent but integrated processes, attachment to the substrate and metamorphosis to a primary polyp. The decoupling of these two settlement processes has been observed in larvae from the coral Acropora. Indeed, the biofilm-derived molecules TBP and GLW-amide neuropeptides induce high levels of larval metamorphosis, but only partial attachment to the substrate (Iwao et al., 2002; Erwin and Szmant, 2010; Tebben et al., 2011; Siboni et al., 2012). The present study showed that QSSM extracts have a converse effect on H. symbiolongicarpus larvae, inducing high rates oflarval attachment, but little or no metamorphosis from an attached disc structure to a primary polyp after 24 hrs. This suggests that a complex network of communication molecules produced by substrate-specific biofilms regulates larval substrate selection and settlement in nature, with some circuits acting on a particular phase of the phenomenon while others on a different one. It has been suggested that larvae from some marine invertebrates such as gastropods and polychaetes have apical cilia that might detect the cues produced by bacterial biofilms (Hadfield, 2011). It is conceivable that such a system operates in H. symbiolongicarpus larvae where QSSMs are sensed at the larva's anterior end, inducing their migration and attachment to the substrate. Once the larva is attached to the substrate by its anterior end, this hypothetical QSSM sensory system might no longer operate, leaving the induction of metamorphosis to another set of cues.
CONCLUSIONS
Relationships between bacteria and marine invertebrates have been evolving for millions of years. Bacteria, due their high metabolic plasticity, have shaped surfaces to create suitable substrates for benthic organisms (Hughes and Sperandio, 2008). QSSMs and/or their target pathways might be added to the list of bacterial-derived cues for larval substrate selection. The information presented in this work may prove relevant in current strategies to repopulate corals on decimated reefs. For example, in the development of coral nurseries, QSSM extracts could be applied to the artificial substrata in order to increase the rate of migration and settlement of coral larvae. These findings also provide new opportunities to understand the cell and molecular mechanisms responsible for larval substrate selection, attachment and metamorphosis.















