INTRODUCTION
Hepatitis E is considered the main cause of viral acute liver failure in developing countries, and nowadays, industrialized countries had been affected by autochthonous infections (Donnelly et al., 2017). Hepatitis E is caused by the hepatitis E Virus (HEV), a non-envelope, positive sense, single strain RNA virus, and possessed three open reading frames: ORF1, ORF2 and ORF3, coding for viral polymerase, capsid and multifunctional small phosphoprotein, respectively (Rogée et al., 2013), and has five known genotypes affecting humans (genotypes 1 and 2) or both humans and animals (genotypes 3, 4 and 7) (Sridhar et al., 2017). HEV infection has been classified as a food-borne zoonosis and the main transmission pathway is through consumption of contaminated water or contaminated food, particularly undercooked or raw pork meat (Rogée et al., 2013; Donnelly et al., 2017).
Antiviral treatment is non-specific, and ribavirin has become the first-line drug for both, acute and chronic HEV infection because the combined therapy with pegylated interferon is contraindicated in immunosuppressed patients (Debing et al., 2014). Resistance to ribavirin has been reported and consequently treatment failure; additionally, anemia is one of the side effects of ribavirin therapy (Donnelly et al., 2017), which is of particular concern for vulnerable patients (pregnant women and immunocompromised), for whom ribavirin could be toxic; therefore, effective and safer antiviral agents against HEV are needed (Hui et al., 2016). However, without a proper in vitro culture system for virus replication, the search for new antiviral compounds is limited.
Hepatitis E Virus life cycle is poorly understood due to the absence of an effective cell culture system (Rogée et al., 2013), but some attempts to propagate the virus in vitro have been done. Primary hepatocytes cultures and pluripotent stem cell (PSC)-derived hepatocytes have been used as effective systems to viral propagation (Helsen et al., 2016; Zhang and Wang, 2016); however, both primary and stem cells are challenging to obtain and to maintain. Continuous cell lines from different origins such as human hepatoma, colon adenocarcinoma, and lung carcinoma had been used (Zhang and Wang, 2016), but high replication efficiency has not been achieved, and cDNA infectious clones had been used to obtain success in vitro infections (Shukla et al., 2012; Qi et al., 2015; Johne et al., 2016). In spite that human lung carcinoma A549 is a non-hepatic cell line, it is the model most used to sustain HEV in vitro infection, and it has been proved that this cell line supports replication of genotypes 1 and 3 (Li et al., 2016), and also a persistently HEV-infected A549 cell line has been obtained (Johne et al., 2016). The aim of this study was to establish an in vitro model of HEV infection for antiviral assays, To this, the A549 cell line was used to isolate and to propagate HEV obtained from a human feces sample of a patient with acute hepatitis, A549 infection with isolated virus was assessed and presence of viral RNA and capsid antigen were detected into the cells after 96 hours post-infection, confirming that HEV recovered from the cell lysate monolayers was infectious.
MATERIALS AND METHODS
Cell culture and virus
Human lung carcinoma A549 cells were obtained from the American Type Culture Collection (ATCC® CCL-185TM) and were grown at 37 °C with 5 % CO2 atmosphere in Dulbecco's modified Eagle's maintenance medium (DMEM, 100 U/mL penicillin, 100 mg/mL streptomycin and 5 % fetal bovine serum). The fecal sample was kindly donated by Servicio de Trasplante Hepático del Hospital Central de las Fuerzas Armadas, Montevideo, Uruguay, and it was obtained from a human feces sample of a South American patient in the acute phase. The sample was previously characterized extracting RNA (TRIzol, LifeTechnologies) from 1 mL of a 10 % suspension of fecal material in phosphate-buffered saline (PBS, Gibco® No Calcium, No Magnesium). To detect the viral genome, an RT-PCR was performed with the 5' end of ORF1 as the target, according to Mirazo et al., (2013). For genotyping, a phylogenetic analysis was implemented using the Maximum-Likelihood method and the Tamura-Nei evolutionary model, using MEGA v6 (Mainardi et al., 2018, submitted).
Virus isolation
For virus isolation, 50 - 70 % -confluent A549 cell monolayers grown in six-well plates (Falcon®) were washed three times with 1 mL PBS. The fecal sample was diluted 2-fold in PBS and filtered through acrodisc syringe filters with a pore size of 0.22 μm (Millex-GV; Millipore Corp., Bedford, MA). A549 monolayers were infected with 0.5 mL of the filtrated sample containing 5.85 x 105 RNA copies/µL per well at 37 °C and 5 % CO2 atmosphere. After two hours the inoculum was removed, cells were washed three times with PBS, and then 2.5 mL of cell maintenance medium was added to each well and incubated at 37 °C in a humidified 5 % CO2 atmosphere. Four days post-infection (dpi) cells were passaged for the first time. Cells were washed, dispersed with 0.25 % trypsin (500 μL), diluted 4-fold in fresh maintenance medium, and 1 mL was added to a new cell well. Infected cells were thus passaged serially and tested by RT-PCR every 3 - 5 days for 41 days. The infected cell cultures were examined daily under an inverted microscope (Nikon TS 100 F), but no specific cytopathic effect was observed. Viral stocks were obtained from infected monolayers by two freeze-thaw cycles in liquid nitrogen, and viruses were quantified by determining the viral genome copy number by RT-qPCR and stored at -80 °C until use.
Viral RNA extraction and quantification
For total RNA isolation, TRIzol (Invitrogen®) was used according to the manufacturer's directions. Then, RT-PCR was performed targeting a region within the ORF1 (partial methyltransferase gene and hypervariable region) as previously described (Mirazo et al., 2013). For RT-qPCR we used the forward (JVHEVF; 5'-GGTGGTTTCTGGGGTGAC-3') and reverse primers (JVHEVR; 5'-AGGGGTTGGTTGGATGAA-3') targeting a region within the ORF3 previously reported (Jothikumar et al., 2006), and the 25 μL mixtures were incubated at 42 °C for 30 minutes, followed by 95 °C for 10 minutes, and 39 cycles at 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 45 s. A melting curve was run to verify the specificity of the products and was performed at 95 °C for 15 s, 60 °C for 15 s and 95 °C for 15 s, collecting data every 0.2 s. A 10-fold serial dilution of the RNA standards (102108 copies) was used for the quantitation of viral genome copies numbers (Mirazo et al., 2018). The RT-qPCR analysis was performed in the ABI 7500 Real-Time PCR System. All procedures were performed by triplicate and data are expressed as means (±S.D.).
Infection assay
To test if viral stock obtained from infected monolayers by two freeze-thaw cycles in liquid nitrogen could be able to infect fresh monolayers, 1 x 105 A549 cells were seeded in 24 well plates (Falcon®), 24 hours before viral infection. Then, cells were treated during two hours at 37 °C with two different inoculum concentrations obtained from cell lysates prepared in serum-free media (1 x 104 or 1 x 105 RNA copies/µL per well, by duplicate in two independent experiments). After that, the viral inoculum was removed and 500 μL of maintenance medium was added to each well. 96 hours post-infection (hpi), viral RNA and antigen detection were done by RT-qPCR and immunofluorescence assays (IFA), respectively. For IFA, cells were fixed with 200 μL per well of 75 % ethanol for 15 minutes; then cells were washed twice with PBS and treated with 200 μL of 0.1 % triton X-100 prepared in PBS. After blocking non-specific binding sites with 1 % bovine serum albumin (BSA) diluted in PBS, a 1:200 dilution of commercial monoclonal antibody against capsid antigen was added (ABCam, UK). Antibody was incubated for one hour at 37 °C and then washed twice in BSA-PBS prior incubation with FITC-labelled secondary antibody (goat anti-mouse IgG ABCam) for one hour at 37 °C. After incubation with the secondary antibody, cells were washed and DAPI diluted 1:100 in PBS was added for five minutes. Cells were rewashed and examined for the presence of HEV antigen by using an inverted fluorescence microscope (Olympus IX81, filters U-MNUA2, 360 - 370 nm, and U-MNIBA3 470 - 495 nm).
RESULTS
Detection of HEV RNA in infected cells monolayers
HEV positive A549 cell cultures were found by RT-PCR after 26 dpi with serial passages every three to four days (Figure 1a). The viral concentration of each day of sampling was quantified. The highest RNA copies number was found after 26 dpi (2 x 106 RNA copies/µL) and viral RNA concentration decrease after day 35. No specific cytopathic effect was observed, and the evaluation finished on day 41 (Figure 1b). Capsid antigen and HEV RNA was detected in A549 cells infected with the isolated virus HEV is a highly tricky virus to isolate and for this reason, we tested if the viral stock isolated after 26 dpi were infectious to new A549 cells. Cells were treated with two viral concentrations prepared in serum-free media (1 x 104 and 1 x 105 copies/µL), and after 96 hpi, capsid antigen was detected perinuclearly and forming foci on A549 cells infected with 1 x 104 copies/µL (Figure 2a) and viral RNA was detected in cell monolayers but no in supernatants. Of note, no statistically significative difference in viral genome copy number between the two viral concentrations used was found (p = 0.68 Mann-Whitney. U-test) (Figure 2b), indicating that this dilution was infectious.
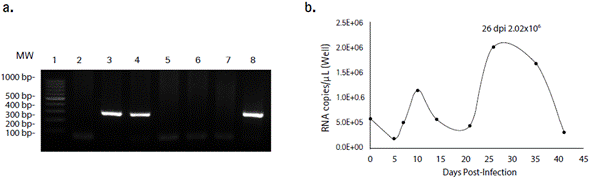
Figure 1 Human feces diluted in PBS were used to isolate HEV in A549 cell line. a. HEV RNA was detected by RT-PCR of a 287 bp ORF1 partial sequence. Viral RNA was detected in cell monolayers after 26 (lane 3) and 41 (lane 4) days post-infection. RNA extracted directly from PBS diluted feces was used as a positive control (lane 8). Non-infected cell cultures (lanes 2, 5 and 6) were tested as a control. Water was used as RT-PCR negative control (lane 7). Thermo Scientific™ GeneRuler™ 100 bp DNA Ladder (lane 1). b. Highest genome copy number of intracellular HEV was achieved at 26 days post-infection and decreased in the subsequent days.
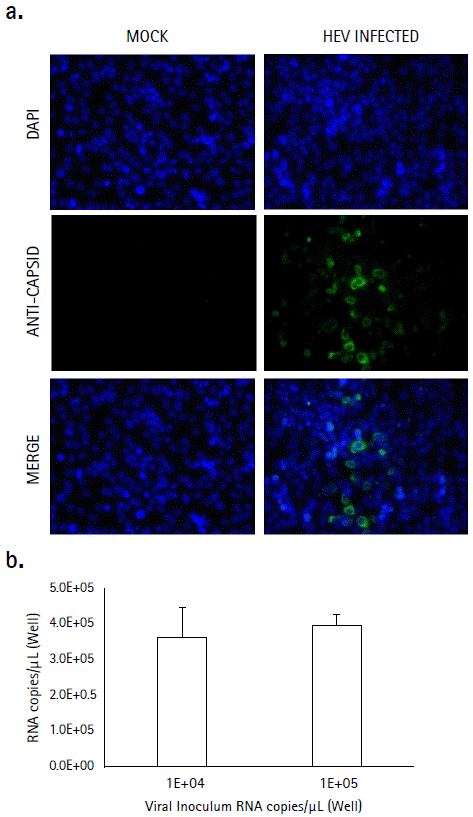
Figure 2 Infection assay. A549 cells were inoculated with 1 x 104 or 1 x 105 copies/µL of the 26 dpi viral stock during 96 hours. a. HEV capsid antigen (ORF-2) was detected in infected cell cultures by indirect immunofluorescence using a monoclonal antibody, followed by a FITC conjugated secondary antibody (green). Nuclei were stained with DAPI (blue). Non-infected cell cultures (mock) were used as a control. b. Viral RNA was detected at 96 hpi, and no statistically difference were found between the two inocula (p = 0.68, Mann-Whitney. U-test). The values are representative of two independent experiments with two replicates each (n = 4). Error bars represent the standard deviation (SD)
DISCUSSION
Hepatitis E virus life cycle is not well understood due to the difficulty to obtain a productive viral infection in vitro. Besides, the few available systems are cell and virus dependent, and in the last 10 years only four cell lines without previous exposure to HEV were found suitable for viral production (Von Nordheim et al., 2016; Zhang and Wang, 2016), and recently it was reported the isolation of wild type strains with high replicative capacity in vitro (Schemmerer et al., 2019).
A549, HEPG2/C3A, PLC/PRF/5, Huh-7 Lunet BLR and MRC-5 cell lines support HEV infection with the virus strain 47 832c of genotype 3, but the higher virus productivity was detected in A549 cell line with 7.3 x 107 copies/mL after 30 days post-infection (Takahashi et al., 2012). For the viral strain JE03-1760F also belonging to genotype 3, viral loads reached 1 x 108 copies/mL, but after 52 days after inoculation (Lorenzo et al., 2008). Under different culture conditions, differences in cell line phenotypes tend to appear, and this could explain why the highest viral genome copy detection in our model was achieved after 26 dpi, in contrast to the other models. Of note, in most of the available cell models, viral recovery had been done from the supernatant (Meister et al., 2019), in contrast to the model used in this study in which viral recovery was obtained from cell monolayers; however, in the study using A549 cells, viral recovery was achieved after 30 days of culturing and the sample was obtained from a human patient serum (Takahashi et al., 2010).
According to this, origin and type of sample is another crucial factor for HEV isolation. It has been reported that genotypes 1, 3 and 4 isolated from human's feces have been successfully propagated and detected between three to 26 days after infecting the cells with at least 1.0 x 104 copies/ mL (Zhang and Wang, 2016). Similarly, the sample we used belonged to genotype 3 and was obtained from human feces (Mainardi et al., 2018, submitted). Animal sample sources as serum, liver and, feces have been reported as adequate for genotype 3 viral isolation, releasing viral progeny to the cell supernatant with 7.3 x 107 RNA copies/mL after 30 dpi (Takahashi et al., 2012). In sharp contrast, we could not detect viral RNA in the supernatant, but after 26 dpi, viral RNA into the cells reached 2 x 106 copies/µL (Figure 1b).
Ferret (Mustela putorius) HEV, another member of the Hepeviridae family, has been used to understand the viral replication cycle. In this case, stool suspension was inoculated in PLC/PRF/5 cells, and viral particles were found associated with cell membranes, and in agreement with our findings, the capsid antigen was in proximity to the nucleus.
Similarly, 1 x 106 RNA copies/mL were found after 32 days post-inoculation (Li et al., 2016).
Takahashi etal., (2012) demonstrated that HEV viral progeny obtained from A549 infected cells with viral particle amounts up to 2 x 104 genome copies/well could support efficient multiplication of HEV in A549 unexposed cells, reaching 7.3 x 107 copies/mL after 30 dpi. Our HEV strain was also able to infect A549 cells and both viral RNA and capsid antigen were detected after 96 hpi when new cell cultures were inoculated with 1 x 104 RNA copies (Figure 2b). Like our model, cell supernatants derived from PSC-hepatocytes were used to infect HepG3/C3A and 2 x 105 RNA copies/mL of intracellular viral RNA was detected after 12 dpi (Helsen et al., 2016).
CONCLUSIONS
Understanding the biology of the HEV life cycle without a proper cell culture system is a difficult task, and therefore, antiviral drug design is still a challenge, despite searching for new antiviral molecules against HEV is a matter of significant interest in the scientific community. Here we report the isolation of HEV genotype 3 on A549 cells with genome detection after serial passages performed every 4 days. The isolated obtained after 26 dpi, was also able to infect non previously exposed A549 cell, and genome and viral protein detection after 96 hours were achieved. This viral isolate could be a helpful tool to study the HEV life cycle and could be used for antiviral drug evaluation.















