INTRODUCTION
The human cytomegalovirus (HCMV) is an enveloped double stranded DNA virus belonging to the family Herpesviridae family, characterized by a very slow viral replication cycle, lifelong latency, and a broad tissue and cell tropism which includes the epithelium, endothelium, monocytes/macrophages, acinar cells and fibroblasts (Vanarsdall et al., 2008). Immunocompetent individuals do not present clinical signs or symptoms during primary infection or reactivation but, in pregnant women, HCMV could induce fetal infection leading to congenital alterations arising from neurological sequelae such as loss of sensorineural hearing, which is its most frequently observed manifestation (Britt, 2017). In immunocompromised patients, HCMV infection is the cause ofsevere complications such as pneumonia, retinitis, gastroenteritis and hepatitis (Griffiths et al., 2015). Currently, the most affected group is that of the patients immunosuppressed after solid or hematopoietic stem cell transplantation and those infected by the human immunodeficiency virus (Ban, 2014).
The HCMV is transmitted via the saliva, urine, breastmilk, genital secretions and transplanted organs (Hamprecht et al., 2008). After primary infection, the HCMV demonstrates latency in the early myeloid CD34+ cell lineage and persists in the granulocyte/macrophage precursors and CD14+ monocytes after cell renovation or differentiation (Requião-Moura et al., 2015). This results in their activation and differentiation that favors transit to the tissues and organs. This phenomenon of macrophage differentiation involves viral replication (reactivation) that is controlled by the immune system (Elder et al., 2019). During immunosuppressive conditions, the viral reactivation is frequent, leading to a lytic infectious phase and resulting clinical manifestations (Lancini et al., 2014).
Depending on the age at which primoinfection and reactivation take place, the HCMV virions can be detected in different bodily fluids such as saliva, urine and genital secretions. In adult infections, congenital infections or infection during the early childhood, the virus can be detected in the urine for months (Twite et al., 2014).
HCMV is diagnosed by the detection of IgM and/or IgG in the serum and, in patients with a probability of reactivation, a viral load test is recommended (Razonable and Hayden, 2013). In both clinical and basic research, virus isolation is a valuable tool but requires multiple sampling and expensive technical protocols. Hence, it is a challenge to investigate the cases of infection. In research laboratories, it is common to use adapted strains of HCMV which have undergone multiple cell culture passages. For example, the AD169 strain, which was obtained from the adenoid tissue of a child and adapted after 50 fibroblast cell passages, or the Towne strain, which was isolated from a patient with congenital infection and passaged 128 times in the human fibroblast WI-38 cells (Prichard etal., 2001). These HCMV strains changed their genetic information after successive passaging along with their tropism and pathogenicity features (Stanton et al., 2010). Other laboratory strains of HCMV have also adapted by low number passaging and retained attributes resembling those of the wild type viruses. For instance, the Merlin strain was isolated from a urine sample and found to adapt after three fibroblast passages prior to its whole genome determination, which facilitated the adoption of this strain as an HCMV standard by WHO (Wilkinson et al., 2015). Having in mind the crucial role ofviral strain obtained from clinical isolates for in vitro research, this work was aimed to isolate and culture HCMV from bodily fluids such as urine, bronchoalveolar lavage, saliva, and plasma samples of pediatric patients with probable or confirmed infection.
MATERIALS AND METHODS
Clinical samples and reference viruses
Bodily fluids to viral isolation attempts were obtained from two patient groups. Firstly, salivary samples were taken from 20 immunosuppressed patients from the Hematopoietic Stem Cell Transplant Unit of the HOMI "Fundación Hospital Pediátrico La Misericordia" in Bogotá, Colombia. The salivary samples were collected once a week during the hospitalization period and a total of 105 samples were obtained. The second source of bodily fluids were hospitalized patients suffering different systemic conditions such as cancer, CMV congenital infection or pneumonia whereby a HCMV infection or reactivation were suspected and a viral load test was prescribed. In total, six urine samples (U), 37 plasma samples (P) and 27 bronchoalveolar lavages (BAL) were collected. All the patients and their guardians signed the informed consent form before the study was initiated and it was approved by the Facultad de Odontología of the Universidad Nacional de Colombia Ethics Committee (Number 11-15, CIE 174 15) and the Fundación Hospital de la Misericordia Ethics Committee (Number 015, CEI 52-18).
The samples were collected in a 15 mL plastic tube containing the culture medium supplemented with penicillin (200 IU/mL) and streptomycin (200 μg/mL). It was transported to the laboratory under controlled temperature conditions of 4°C, centrifuged (4,500 rpm, 10 min, 4 °C), and the supernatants were stored at -80 °C until use. The Towne strain virus was obtained from the lab in VIREM, Universidad del Valle, Cali, Colombia (Dr. Beatriz Parra, February 2018) and the Merlin strain virus was bought from the UK Repository (NCVP0302163v).
Isolation and harvesting of the virus
The MRC-5 human lung embryonic fibroblast cell line (ATCC® CCL-171, United Stated) was used to isolate the viruses from the samples. The Cells were seeded on 24-well plates at a density of 1X105 cells/well and maintained in DMEM and 10 % fetal bovine serum (FBS) at 37 °C in an incubator with 5 % CO2 for 24 h. Saliva and BAL were directly inoculated without diluting (330 μL) in triplicates, while those of the urine and plasma were diluted in the culture medium to 1:2 and 1:20, respectively. All the samples were processed as per the modified shell vial protocol. The cell-inoculum plates were centrifuged at 1000 g for 30 min and incubated further for two hours at 37 °C before addition of 330 μL of culture medium with a 4 % FBS supplement and an antibiotic (2X). The culture was maintained either for two weeks or until CPE was observed. This culture stage was called the zero passage (P0).
Supernatants of the inoculated cells (500 μL) were re-inoculated in P25 culture flasks and rocked gently for 1 h at room temperature. Fresh medium was added to the flasks and incubated either for two more weeks or until a CPE was observed in at least 80 % of the monolayer. Here, the cells were scraped and sonicated (4 cycles, 30 seconds and 40 % amplitude) to obtain the virus and further harvesting from P1 to P3 was carried out. The harvested viral cultures were then cryopreserved at -80 °C in FBS at a concentration of 10 %.
Virus titration
Strains of the virus and the harvested viruses were titrated using the tissue culture infectious dose 50 (TCID50) in primary human gingival fibroblasts. Approximately 2>104 cells were seeded in each well of the 96-well plates. 24 h later, they were inoculated with a 10-fold serial dilution of the different HCMV strains and incubated for 2 h. Six wells were inoculated per dilution. After 2 hours, the inoculum was discarded, and fresh medium was added. Inverted phase contrast microscopy was used to detect CPE in the wells after 48 h post infection. Titers of the virus were calculated using the Spearman-Karber algorithm.
Polymerase Chain Reaction (PCR), virus detection and confirmation
DNA and RNA were isolated from the supernatants and samples of the viral harvests (Stratec Cat. 1040, Germany) in order to conduct amplification and evaluation as per the methods described in the protocols (Table 1) for conventional PCR.
RESULTS
Although 29 of the 105 salivary samples that were analyzed were found to be positive for HCMV (27.6 %), inoculation of all the samples was done on MRC-5 cells. CPE was observed in 61 samples (Fig. 1a), of which, majority demonstrated the effects during the first 48 h post inoculation. The phase contrast observation enabled us to describe different types of CPE ranging from large, unique, round, refringent cells and cell clustering with no membrane fusion to cell clustering with membrane fusion (syncytia). Like those inoculated with the Towne strain, these cell clusters appeared on top of the monolayer. From the cultures demonstrating CPE positively, supernatants of 21 were positive for HCMV, as determined by the PCR results, but majority of them were also found to be positive for the Herpes Simplex Virus (HSV), which could explain the early onset of CPE. Positive results for HSV-1 or HSV-2 were also obtained in the seven isolates that were originally negative for HSV but later passaged sequentially up to P3 to reconfirm on the presence HCMV (Fig. 1b).
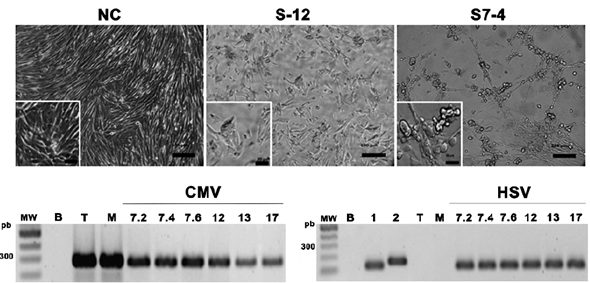
Figure 1 HCMV Isolation and characterization from saliva samples. Photomicrography of culture during viral isolation (P0) on uninfected MRC-5 (NC negative control), Inoculation with patient numbers 12 and 7 saliva samples (S12, S7), and S7.4 is saliva taken during the fourth week of hospitalization. Bar correspond to 100 mm and 50 mm in the left corner insets. PCR detection of HCMV and HSV in viral harvests P3. MW (Molecular weight marker), B (PCR negative control), T (Reference strain: Towne), M (Reference strain: Merlin), HSV1 y HSV2 (Positive controls for each HSV type).
In order to isolate the virus, when plasma samples positive for HCMV were inoculated, coagulation was induced immediately on the cell monolayer along with cell detachment. Hence, plasma samples were not used for the process of virus isolation. Two urine samples were positive for HCMV and inoculated into the virus culture. The sample U24 induced an abnormal CPE and showed the presence of fusiform cells at 24 h post inoculation but large clusters of rounds and refringent cells after 48 h (Fig. 2a). The PCR for HCMV was positive in these supernatants and negative for adventitious viruses such as adenoviruses (AdV) and enteroviruses (EV) (Fig. 2b). Since the second urine sample (U17) positive for HCMV induced a slight CPE and the P0 supernatants were found to be HCMV positive, successive passages still P3 were carried out which were also HCMV but did not induce CPE.
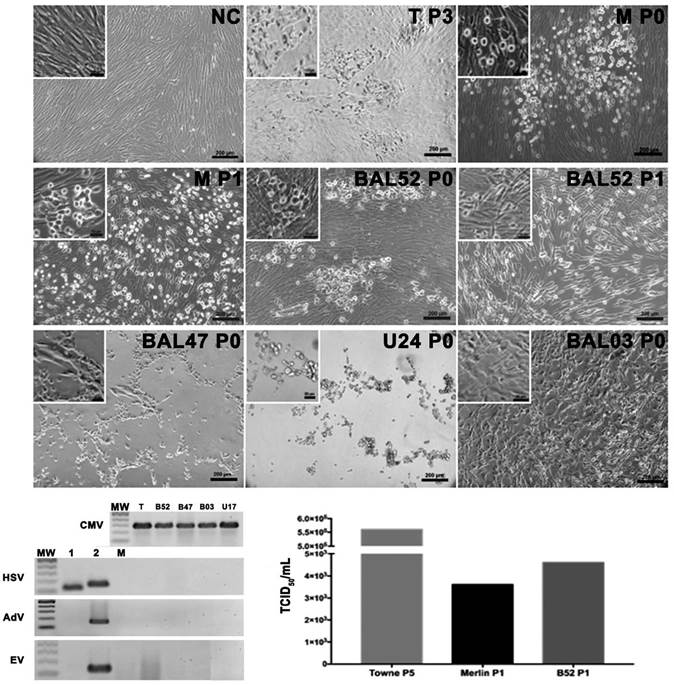
Figure 2 HCMV isolation from bronchoalveolar lavage and urine samples. Photomicrography of Merlin strain (P0) and B52 after 10 days post inoculation. Early aspect (24 or 48 h p.i.) of cell cultures after B47, U24 and B03 initial (P0) sample inoculation. Second passage inoculation (P1) cell culture aspect after 10 days p.i. Negative control (NC) uninfected MRC-5 culture. PCR products for HCMV in each isolate, HSV showing the positive controls for HSV type 1 and 2, RT-PCR for EV and PCR for AdV with the positive control for each one. MW (Molecular weight marker), C (PCR Negative Control), T (Reference strain: Towne), M (Reference strain: Merlin), B52 (BAL52) and B47 (BAL47). c. Virus titration by TCID of the reference strains and B52 sample.
On the other hand, seven BAL samples positive for HCMV were inoculated for isolation of the virus, of which, three induced CPE with variable morphologies. For instance, the sample BAL03 induced CPE at 48 h post inoculation and demonstrated a fusiform cell morphology (Fig. 2a), while that of BAL47 was characterized by round refringent cells only in the P0, a finding like that described above for the U24 sample. Furthermore, it was observed that CPE was induced in the BAL52 sample 10 days after inoculation, the morphology of which was like that of the inoculated Towne or Merlin strains. In brief, it was observed that cells appeared as foci with a round and smooth membrane that later extended in different directions, enlarged in size and changes occurred in the nucleus until final detachment. For this strain, during adaptation in P1, the CPE was close to that of the Merlin strain in which infected cell clusters had spread occupying the entire monolayer and demonstrated round refringent that is classic of its CPE. Thus, this adapted strain did not belong to the group of adventitious viruses, evidently induced a CPE similar to that of the reference strains when titrated and yielded a titer (4.6>103 TCID50/mL) similar to that of the Merlin strain (3.6>103 TCID50/mL) but lower compared to that of the Towne strain (5.6>105 TCID50/mL) (Fig. 2c).
DISCUSSION
This study confirmed that isolation of the HCMV is intensive and arduous and requires a strict selection of the bodily fluids to begin with. This can be elucidated from the fact that all saliva samples were positive for HSV at the beginning or after three passages, which could be explained by the extensive prevalence of these viruses in developing countries where 90 % and 50 % of people are positive for HSV-1 and HSV-2 respectively (Paz-Bailey et al., 2008), in addition to the salivary shedding demonstrated in transplant recipients (Bohórquez et al., 2016) or in asymptomatic individuals (Kauffman et al., 2005). These saliva samples induced an early CPE (48 h) with a morphological pattern different from that of the reference strains, which consisted of round and refringent cells distributed unevenly on top of the monolayer similar to those reported previously in Hep-2 cells where HSV induced syncytia were observed even at 24 h after inoculation (Nozawa et al., 2014).
Isolation of the HCMV has always been a challenge since the time when first attempts were made by Gregory and Menegus (1983) using MRC-5 and WI-38 cells to collect the supernatants 17- and 24-days post inoculation. Thereafter, a protocol for inoculation and centrifugation (shell vial) was described by Leland et al, in 1989, the application of which resulted in shorter periods of observation and CPE manifestation. Here, in addition to the shell vial technique, a definite inoculum was maintained which enabled the CPE to appear at day 10 post inoculation in both, the diluted reference Merlin strain and the BAL samples. It can be elucidated that the reference Towne strain of HCMV showed adaptation to the culture because the characteristic CPE was induced early and easily.
After the evaluation of 175 samples of bodily fluid, the sample BAL52 demonstrated the classic morphological pattern reported for HCMV (Leland and Ginocchio, 2007) with an appearance like that of the reference strains. Due to the slow viral cycle (48 h), the CPE appear after 10 days of inoculation in the form of foci in large round cells with cluster aggregation (Hematian et al., 2016). We discarded the possibility of the presence of adventitious viruses such as the adenoviruses or enteroviruses which are frequently shed in the lung fluids of children (Jain et al., 2015; Montes et al., 2019), thereby confirming that the observed CPE after the inoculation of BAL52 was not related to the effects of the adventitious viruses but with that of the HCMV.
Urine is the main bodily fluid from where the HCMV is excreted in children below the age of 5 years or by neonates during congenital infections (Vestergaard et al., 2018). Six patient samples were evaluated using PCR and two were found to be positive for HCMV positive by PCR. However, they exhibited an atypical CPE after the first inoculation (P0) and did not appear in the subsequent passages. It is presumable that in addition to HCMV, the samples are likely to contain the polyomavirus as well, which has a high prevalence (65 to 90 %) and is frequently excreted in the urine of pediatric patients (Knowles, 2006). Although this virus was not isolated from the urine samples, the BK polyoma virus infects the bladder fibroblasts and induces a CPE that gives individual round refringent cells with a complete damage of the monolayer (An et al., 2019) resembling to that observed after inoculation with the U17 sample.
Being a pathogen, the HCMV is involved in many pathologies and complications in immunosuppressed individuals. Virus strains that have been properly characterized are a necessary tool for understanding their pathogenesis and relationship with the immune system.
Thus, attempts to establish more laboratory strains is a key task for virologists.
CONCLUSIONS
A modified shell vial protocol was used for the isolation of HCMV and to successfully establish a new low passage strain from BAL. The presence of other viruses inducing CPE was also evaluated but differences in cytology led to the discarding of this supposition. The use of bodily fluids such urine, saliva and plasma are not suitable for the isolation of HCMV, but bronchoalveolar lavage is a potential candidate because these samples are collected from sites deep within the bronchi and alveoli which are less prone to contamination by other viruses, making these samples more appropriate for the isolation of HCMV.