INTRODUCCIÓN
Semliki Forest Virus (SFV) is a member of the Alphavirus genus, family Togaviridae. The alphaviruses replication strategy is highly efficient; moreover, SFV has a wide host range from insects to mammals and its genome insertion capacity has been used for the development of a suitable expression system of heterologous proteins in eukaryotic cells (Henao et al,. 2007). The SFV system is based on a cDNA copy of the SFV genome with a deletion of the structural genes to allow the insertion of a heterologous gene. The recombinant SFV particles are obtained by co-transfection with three different RNAs. The first one corresponding to the replicon (coding four nonstructural proteins of SFV or transcriptase complex) and the sequence of the heterologous gene, the second one is the sequence coding for SFV capsid protein, and the third one, the sequence coding for SFV envelope proteins.
The recombinant viral particles contain a defective genome, which leads to RNA replication and expression of the replication complex and the heterologous protein. However, no infection occurs due to the absence of the alphavirus structural genes, therefore, providing a system with a high biosafety level (Berglund et al., 1999). The SFV vector has been used for heterologous protein expression in different types of mammalian cells, also for vaccine production, and gene therapy applications (Vidalin et al., 2000; Lemmonier et al., 2002; Hourioux et al., 2007; Ip et al., 2014). Therefore, it could be a useful tool to study viral proteins including the properties of the HCV proteins.
The HCV is an RNA virus from the Flaviviridae family, the Hepacivirus genus. The viral genome is a positive single-stranded RNA of 9.6 kb that encodes a polyprotein of approximately 3000 amino acids. Cellular and viral proteases process the polyprotein into structural proteins (Core, E1, and E2) and nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Lin, 2006; Welbourn and Pause, 2006).
The World Health Organization (WHO) estimates 71 million cases of chronic hepatitis C infection around the world (WHO, 2018). Indeed, between 60 to 80 % of the patients infected with HCV develop chronic infection, and 15-30 % of them evolve to end-stage liver diseases such as cirrhosis and Hepatocellular Carcinoma (HCC) (WHO et al., 2017; WHO, 2018). The HCC is the most common type of primary liver cancer and the fourth most common cause of cancer-related death (WHO and International Agency for Research on Cancer (IARC), 2018).
The first HCV treatment option was Interferon type I with unsuccessful results in more than 50 % of patients infected with HCV genotype 1, the viral genotype with the highest distribution worldwide. However, the recent developed direct-acting antivirals (DAA) have better outcomes with a 95 % cure rate (González-Grande et al., 2016). Unfortunately, the access to this treatment is difficult due to the high cost (Rosenthal and Graham, 2016), and the viral clearance after the DAA treatment is not associated with HCC risk reduction in all cases (Conti et al., 2016).
The HCV infection causes chronic inflammation, and the viral proteins induce oxidative stress leading to fibrosis and cirrhosis (Sukowati, 2016). Oxidative stress occurs with the excessive production of reactive oxygen species (ROS), and long term oxidative stress induces DNA damage (Sukowati, 2016). Besides, Core, NS3, and NS5A HCV proteins have shown oncogenic activity related to pathways dysregulation that promotes malignant transformation of hepatocytes morphology, contributing to hepatocarcinogenesis (Akkari et al., 2012; IARC, 2012; Vescovo et al., 2016).
HCV Core protein is related to some oncogenic processes, including effects on the tumor suppressor protein p53. This cellular protein is a transcription factor that regulates the cell cycle, programmed cell death, response to cell stress and DNA repair. The Core protein could induce mutations in p53 gene like G to A, and the mirror transition C to T affecting p53 functions (Kao et al., 2004; McGivern and Lemon, 2011; Tornesello et al., 2013; Poole et al., 2018).
HCV Core protein could also enhance the expression of the cyclin-dependent kinase inhibitor p21 waf1/CIP1 , which is a target of p53 and regulates activities such as cell-cycle and tumor formation through a p53-dependent manner, in the human hepatoma (Lu et al., 1999; Kwun and Jang, 2003; Feng et al., 2015). Other studies indicate that HCV Core protein could repress transcription of the p21 gene through the tumor growth factor β (TGF-β) pathway, as demonstrated by in vitro transient expression assays using Huh7, murine fibroblasts (NIH 3T3) and primary hepatocytes isolated from transgenic mice (Lee et al., 2002; Kwun and Jang, 2003; Jahan et al., 2011).
Since HCV is essentially a hepatotropic virus, the expression of viral proteins in cells with the hepatic environment is a model to study the properties of the HCV proteins. Since the HCV replication system in vitro has limitations, then the expression of viral proteins using viral vectors could be an appropriate model to study the properties of the HCV Core protein in a hepatoma human cell line (Bartenschlager and Lohmann, 2000). Consequently, the present study aimed to determine the efficiency of the SFV expression system in the HepG2 human liver cell line estimating the expression of the Green Fluorescent Protein (GFP) and HCV Core protein. Moreover, we evaluated the effect of transitory HCV Core protein expression on p53 mRNA level in HepG2 cells.
MATERIALS AND METHODS
Cell lines
Human Hepatoma cells HepG2 were maintained in DMEM (Dulbecco's Modified Eagle's Medium, Gibco), supplemented with 10 % heated inactivated fetal bovine serum (FBS) (Gibco), 1 % penicillin/streptomycin (ICN), 1 mM sodium pyruvate (ICN), 25 mM HEPES, 1 % L-Glutamine (ICN) and incubated at 37 °C in a humidified atmosphere with 5 % CO2. BHK-21 cells were maintained in BHK medium (Gibco, Life Technologies, Scotland), supplemented with 10 % tryptose phosphate broth, gentamicin 5 μg/mL (Thermo Fisher Scientific8, USA) and 10 % FBS (Valbiotech, France).
Recombinant viral particles rSFV-GFP and rSFV-Core
The pSFV1-GFP and the pSFV1-Core were constructed using the pSFV-1 plasmid as described previously (Navas et al., 2019). The sequences were obtained by PCR amplification from the pEGFP (GFP, 754 bp) and the infectious clone p90/HCV FL-Long pU (HCV genome, nucleotides 342-914). Then, the sequences were cloned into pSFV-1 at the BamHI site downstream of the 26S viral promoter.
In vitro transcription of RNA from SFV1-GFP, SFV1-Core and pSFV-helpers was carried out using the large-scale mMessage mMachine Sp6 transcription kit (Ambion Inc, Austin, TX) and DNA previously treated with proteinase K and SDS. Then BHK-21 cells were co-transfected with the mRNAs synthesized (SFV1-GFP and SFV-helpers, or SFV1-Core and SFV-helpers) by electroporation (Gene Pulser II, BioRad, Richmond, CA), as previously described (Liljestrom and Garoff, 1993). Culture supernatants of co-transfected cells were harvested 36 hours later.
A serial passage assay was performed in BHK-21 cells to determine the titer of the recombinant SFV (rSFV) particles, rSFV1-GFP, and rSFV1-Core. The proteins expression levels were measured 24 hours post-transduction (h.p.t.) by flow cytometry or indirect immunofluorescence with the human monoclonal anti-Core B12.F8 antibody, which recognizes a major B cell epitope within the amino-terminal region of the HCV Core (Cerino et al., 1993), and a secondary antihuman IgG FITC-conjugated Ab (Sigma, San Louis, MO). The viral titer was calculated using the Reed-Munch Tissue Culture Infectious Dose 50 (TCID50) method.
HepG2 cells transduction with the recombinant viral particles
HepG2 and BHK-21 cells (2 x 106 cells) were transduced with rSFV-GFP or rSFV-Core recombinant viral particles at a multiplicity of infection (MOI) of 0.5, as previously described (Navas et al., 2019). After transduction, cells were incubated in 5 ml offresh DMEM or BHK medium supplemented with 10 % FCS at 37 °C for 24 hours. Respectively as a negative control, we treated cells without recombinant viral particles at the same conditions (mock cells).
We evaluated GFP expression to determine the efficiency of SFV as an expression system in the hepatoma cell line HepG2. The GFP expression level was determined by flow cytometry after the transduction of HepG2 cells with a dilution corresponding to the rSFV1-GFP TCID50. GFP expression was detected in transduced HepG2 cells 24, 48, 72, and 96 h.p.t by flow cytometry and by fluorescence microscopy (Nikon ELWD 0.3-OD75) 24 hours h.p.t.
Expression of the HCV Core protein was evaluated by indirect immunofluorescence using the B12.F8 Ab at the same time points as GFP expression using the dilution corresponding to the rSFV1-Core TCID50. The expression levels of GFP and HCV Core protein in transduced HepG2 cells were also confirmed by western blot.
Immunofluorescence
Cell monolayers were fixed (3 % paraformaldehyde) and then treated with 0.5 % Triton X-100 (Sigma, USA). Detection of the HCV Core protein was performed using the human monoclonal B12.F8 antibody diluted 1:25 in PBS containing 1 % bovine serum albumin (BSA) (Sigma, USA). Followed by the secondary antibody FITC conjugated goat anti-human 1gG1 (Sigma, USA) diluted 1:50 in PBS 1 % BSA and Evans blue.
Western blot
Cell were harvested 24 h.p.t and lysed in this buffer: 50 mM Tris, pH 8.0, 150 mM NaCl, 1 % Nonidet P40, 0.5 % sodium deoxycholate, 0.1 % SDS and 0.5 % protease inhibitor cocktail, and incubated for ten min at 4 oC. The proteins were quantified using the Bradford assay and then loaded on a 10 % sodium dodecyl sulfate (SDS) polyacrylamide gel (PAGE) in reducing conditions assuring equal protein loading, blotted onto a PVDF membrane (Amersham-Pharmacia, Uppsala, Sweden), incubated with the B12. F8 Ab and secondary anti-human IgG HRP-conjugated Ab (Amersham), and revealed by chemiluminescence (ECL Western Blot, Amersham) according to the manufacturer's instructions (Sambrook et al., 1989). The immunoblots were quantified by densitometry (un-scan-it v.6, Silk Scientific, Orem, UT), and protein levels were compared with the signal corresponding to a known concentration of a truncated isoform of Core (120 amino acids)
Total RNA extraction
Total RNA from transduced and mock HepG2 cells was extracted using TRIzol® reagent (Invitrogen life technologies) 24 h.p.t. Cell cultures were lysed directly in a culture dish by adding 1 ml of TRIzol® per 10 cm2 of the culture dish area. Total RNA was diluted 1:100 and quantified by spectrophotometry (Spectronic GENESYS 10 UV, Thermo Electron Corporation), and the absorbance ratio A260/A280 was used to determine the purity of the extraction.
Semi-quantitative rt-PCR of p53 mRNA
RNA, cDNA, and magnesium concentrations and PCR cycles were adjusted to achieve optimal PCR conditions for quantification. RNA (2.66 mg) were reverse transcribed in the presence of 0.5 mg of primer OligodT (Promega), 20 U RNAse inhibitor (Fermentas, Cleveland USA), 200 U Moloney Murine Leukemia Virus Reverse Transcriptase (Fermentas), and 1 mM dNTP's (Promega) in a final volume of 20 mL. One-tenth of cDNA was amplified with Taq polymerase (Fermentas) for p53 amplification (94 °C, 4 min, 35 cycles of 94 °C, 30 s, 55 °C, 30 s, 72 °C, 1 min, 72 °C, 4 min) and glyceraldehyde three phosphate dehydrogenase (GAPDH) amplification (90 °C, 5 min, 22 cycles 90 °C, 1 min, 60 °C, 1 min, 72 °C, 1 min, 72 °C, 10 min). Concentrations of 3 mM MgCl2, 1:2 cDNA dilutions and 200 nM of Inter exon primers 5'-TTGCCGTCCCAAGCAATGGATG-3' (nt 1202712048), 5'-TCCTGACCTGGAGTCTTCCAG-3' (nt 1409314116) were used for p53 amplification. And the primers 5'-CCCTTCATTGACCTCAACTACATGG-3' (nt 3926-3950), 5"AGTCTTCTGGGTGGCAGTGATGG 3' (nt 4953-4975) were used for GAPDH amplification (Kaboev et al., 2000). All the amplification products were run in agarose gel electrophoresis at 1.2 % for 60 minutes at 80 V.
Densitometry analysis
The amplification products obtained from standardization assays and semi-quantitative RT-PCR were analyzed in 1.2 % agarose gel electrophoresis stained with ethidium bromide and quantified by densitometry using the program Image J (V1.4.3 NIH). We performed the analyses following the recommendations described in Current Protocols in Cell Biology (Gerstein, 2001). p53 mRNA relative levels were calculated by measuring the relative ratios (RR) p53/GAPDH acquired from the media of five independent experiments obtained by densitometry analysis of gel bands.
Statistical analysis
The normality of the data was probed before proceeding to the statistical analysis. The analysis of p53 mRNA expression levels was accomplished using the Statgraphics Centurión XV package (StatPoint, Inc 2005). The difference between medians was analyzed by a non-parametric Kruskal and Wallis Test performed using whole densitometry values (p = 0.0000; H = 130.073 > 5.99 (X2 0.05, 2) and p =0.0000; H= 119.536 > 7.81 (X2 0.05, 3)). Additionally, we conducted a one-way ANOVA and a multiple comparison media test (Tukey Test) to analyze differences between p53 mRNA levels in transduced and non-transduced cells. The relative ratios (RR) mRNA p53 /mRNA GAPDH were calculated from media densitometric values of five independent experiments.
RESULTS
GFP expression in transduced-HepG2 cells
The reporter GFP expression was first evaluated to determine the efficiency of SFV as an expression system in the hepatoma cell line HepG2, taking into account that the appearance of GFP has been demonstrated in different cell lines, as being a suitable indicator of the efficiency of the SFV system (Ehrengruber et al., 1999).
The GFP expression level was determined by flow cytometry after HepG2 cells transduction with a dilution corresponding to the rSFV1-GFP TCID50. GFP expression was detected at 24 to 96 h.p.t. in rSFV transduced cells with a peak between 24 and 48 h.p.t. (Fig. 1a). The percentage of cells with GFP expression was similar between rSFV-transduced HepG2 (70.4 %) and BHK-21 cells (71.3 %) 24 h.p.t. (Fig. 1b).
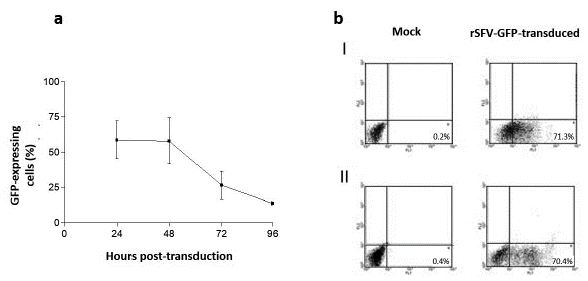
Figure 1 GFP expression in transduced cells using rSFV. a. GFP expression was detected by flow cytometry in rSFV transduced HepG2 cells. The arithmetic means expressions from three different experiments ± SD. b. Dot plot analysis of GFP expression in rSFV transduced cells. I. BHK-21 cells 24 h.p.t. II. HepG2 cells 24 h.p.t. Fluorescent events in region 4 of the channel FL1 correspond to cells expressing GFP. The percentage of fluorescent cells is indicated in each dot plot.
This result indicates the transduction efficiency and the replication of rSFV in this human hepatoma cell line and confirms the data previously published (DiCiommo and Bremner, 1998; Ehrengruber et al., 1999).
HCV core protein expression in HepG2 cells using rSFV
Considering the results obtained with rSFV1-GFP in HepG2, the transduction efficiency of the expression of the HCV Core protein was investigated. Expression of the HCV Core protein was evaluated by indirect immunofluorescence using the B12.F8 Ab at the same time points as GFP expression using the dilution corresponding to the rSFV1-Core TCID50. Even if the GFP expression as a heterologous protein in transduced-HepG2 cells was demonstrated, the HCV Core protein was detected only in 10 to 25 % of cells between 24 and 48 h.p.t., compared to around 60 % of cells positive for GFP expression. The HCV Core expression level was also lower compared with the results obtained in rSFV1-Core-transduced BHK-21 cells (50 % 24 h.p.t.) (Fig. 2a).
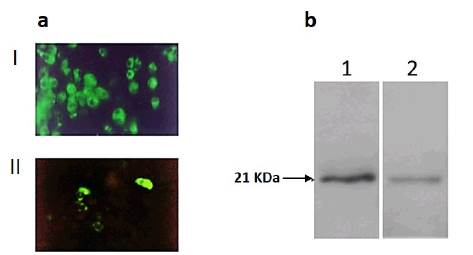
Figure 2 HCV Core protein expression in transduced cells using rSFV. a. Detection of HCV Core expression by indirect immunofluorescence (Magnification 40x) I. BHK-21 cells 24 h.p.t. II. HepG2 cells 24 h.p.t. b. Western blot analysis of HCV Core protein expression in transduced cells with rSFV-Core. Lane 1 BHK-21 cells 24 h.p.t. Lane 2 HepG2 cells 24 h.p.t
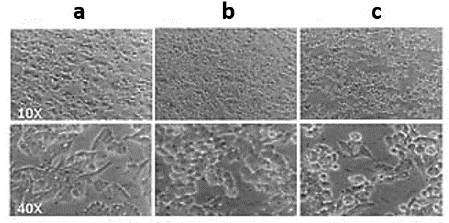
The difference observed between the HCV Core protein expression levels in rSFV transduced cells was also confirmed by western blot. Immunoblotting analysis indicated an 11-fold increase of HCV Core expression level in BHK-21 cells compared to transduced HepG2 cells (Fig. 2b). In addition to the lower efficiency of the recombinant SFV particles for HCV Core expression in HepG2 cells, we observed a marked cytopathic effect (CPE) in rSFV1-Core-transduced HepG2 cells 24 h.p.t. (Fig. 3).
p53 mRNA levels in HepG2 cells transduced with rSFV-GFP and rSFV-core
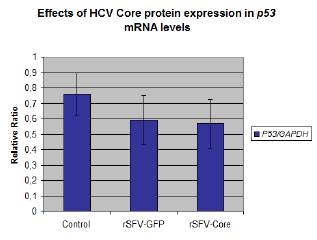
Five independent RT-PCR experiments for p53 mRNA and GAPDH mRNA amplifications were performed to compare mRNA expression levels in transduced-cells transduced, GFP transduced, and control cells. A decrease of p53 mRNA level was observed in transduced cells compare to mock cells. Moreover, the lowest p53 mRNA level was detected in rSFV-Core transduced cells (Fig. 4).
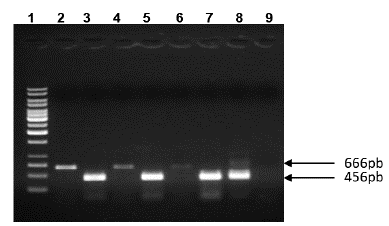
Figure 4 RT-PCR assay for p53 mRNA and GAPDH mRNA level in rSRV transduced-HepG2. Lane 1: Molecular weight 1 kb (Ladder, Fermentas). RT-PCR of p53 mRNA Lanes 2, 4, 6. Lane 2: Mock cells, Lane 4: rSFV-GFP transduced HepG2 cells, Lane 6: rSFV-Core transduced HepG2 cells. RT-PCR of GAPDH mRNA Lanes 3, 5, 7-9. Lane 3: mock HepG2 cells, Lane 5: rSFV-GFP transduced HepG2 cells, Lane 7: rSFV-Core transduced HepG2 cells. Lane 8: Positive control and Lane 9: Negative control. Agarose gel electrophoresis 1 %.
The p53 mRNA/GAPDH mRNA relative ratios obtained from the experiments were analyzed with ANOVA (p = 0.1356). Although the relative ratio was lower in rSFV-Core transduced cells, the data did not show a significant difference between rSFV-GFP and rSFV-Core HepG2 cells (Fig. 5).
DISCUSSION
In this study, we used the recombinant SFV as a viral vector in the hepatoma cell line HepG2, and we performed a semiquantitative RT-PCR for p53 mRNA to investigate the effect of the HCV Core protein in p53 transcription in transduced cells.
The GFP expression levels detected in transduced HepG2 and BHK-21 cells 24 h.p.t are in agreement with the results previously described (DiCiommo and Bremner, 1998; Ehrengruber et al., 1999). Interestingly, the GFP expression in transduced HepG2 cells even at 96 h.p.t, which differs from the results obtained in transduced BHK-21 cells (Fig. 1) and suggest that compared to BHK-21 cells, HepG2 cells are more resistant to SFV replication.
However, according to Wahlfors et al. (2000), the relative transduction efficiency of Sindbis virus on HepG2 cells is very low compared to BHK cells, 293T cells, and human primary fibroblasts, the best targets for both Sindbis virus and SFV. The contradictory results obtained between the present study of high transduction efficiency of SFV on HepG2 cells for GFP expression and the research by Wahlfors et al. (2000) could be due to differences between HepG2 cell clones and probably host range differences between SFV and Sindbis virus.
Considering the transduction efficiency observed in HepG2 using rSFV-GFP, the expression of the HCV Core protein was investigated. In contrast to the GFP expression level, the effectiveness of the SFV system for the HCV Core protein expression in HepG2 cells was three to seven times lower than for GFP.
The low HCV Core expression level described in this study differs from the report of SFV-transduction efficiency in this cell line, where a high level of hepatitis E virus (HEV) capsid protein expression in rSFV-transduced HepG2 cells was described (Torresi et al., 1997). However, the failure to induce anti-Core specific cytotoxic T lymphocytes in mice after intramuscular injection with rSFV particles described by Vidalin et al. (2000), could be due to the low level of expression of the HCV Core in cells, different from BHK using this viral vector, as observed in the present study.
A strategy aimed at increasing the expression level when using the SFV vector system was reported for the HCV NS3 protein by Frelin et al. (2004). The authors demonstrated a significant increase in HCV NS3 expression level by adapting the viral sequence to the most commonly used codons by human cells. This strategy could be envisioned to improve the transduction efficiency of SFV in HepG2 cells for the expression of specific proteins.
This study showed a reduced level of HCV Core protein expression compared with GFP. rSFV1-GFP-transduced HepG2 cells exhibited not only higher heterologous protein expression but also less critical CPE than rSFV1-Core transduced HepG2 cells. Enhanced CPE due to the presence of replication-competent viruses (RCV) in the rSFV1-Core particles collected was kept under control by inoculation of transduced HepG2 cell supernatant in BHK-21 monolayers. The CPE was not detected in these assays suggesting RCVs were absent or present at low levels (data not shown).
The marked CPE could be due to the apoptosis induction by the HCV Core protein, probably associated with the regulation of p53 or p53-related proteins. Core protein affects the p53 protein level and its transcriptional activity (Lu et al., 1999; Kwak et al., 2017). This viral protein could regulate p53 functions via protein-protein interaction, modulating the transcription or with post-translational modifications (Kao et al., 2004). Indeed, Jahan et al. (2011) showed that HCV Core protein from genotype 3a downregulates the expression of p53 mRNA in human hepatoma cell line Huh7. However, in our study, no significant difference in p53 mRNA levels was demonstrated between rSFV-Core compared to rSFV-GFP transduced (Fig 5). Indeed, the limitations of the present study are the low-resolution technique to show differences at the transcription level of mRNA p53 and possibly the variability among the transduction assays.
In the present study, decreased HCV Core protein expression was not related to the SFV replicon because vector efficiency was demonstrated with reporter protein expression. This result could be associated with the induction of oxidative cellular stress by increased reactive oxygen species (ROS) production and subsequent autophagy and viral protein degradation as described in HuH-7 cells expressing HCV Core protein (Rios-Ocampo et al., 2019). The accumulation of the HCV Core protein in the endoplasmic reticulum (ER) leads to ER stress which can result in changes in cell homeostasis as an adaptive response mammalian cells activate the Unfolded Protein Response to reduce the stress decreasing the protein load at the ER (Rios-Ocampo et al., 2019). Moreover, the HCV Core protein can induce toxicity in HepG2 cells by activation of triglycerides biosynthesis and subsequent accumulation of lipids (Hourioux et al., 2007; Kwak et al., 2017).
CONCLUSION
This study confirms that rSFV can be a useful tool for transient heterologous protein expression in human liver cell lines. Further studies are required to demonstrate if the low level HCV Core protein expression described in rSFV transduced-HepG2 cells is related to induction of ER Stress and protein degradation.