INTRODUCTION
Different vertebrate models such as the western clawed frog (Xenopus tropicalis) (Gray, 1864), zebrafish (Danio rerio) (Hamilton-Buchanan, 1822), chicken (Gallus gallus) (Linnaeus, 1758) and the common white mouse (Mus musculus) (Linnaeus, 1758), have been used in the field of developmental biology to understand the molecular and cellular processes that regulate the development of different organisms (Bahr, 2008; Grainger, 2012; Meyers, 2018). Some of the main characteristics that make these organisms very useful in this field of research are that they are easy to breed in laboratory conditions, have high numbers of offspring and different genomic tools are available (Banerjee, 2014; Mueller et al., 2015). In the field of developmental biology, these model facilitate the study of processes such as embryogenesis (Mummery et al., 2011), evolution (Sommer, 2009), aging (Sachs and Buchholz, 2017), tissue regeneration (Brockes and Kumar, 2008; Banerjee, 2014), and metamorphosis (Sachs and Buchholz, 2017). Also, these models are very important in biomedical research because they help to elucidate the molecular pathways that are involved in human diseases, allowing the testing of different substances in vivo (Zon, 2016) to discover new therapeutic strategies in the treatment of complex diseases such as cancer (Sachs and Buchholz, 2017), cardiovascular (Zaragoza et al., 2011), and degenerative diseases (Sachs and Buchholz, 2017).
In Colombia, the main vertebrate models used in developmental biology and regeneration research, include chicken embryos (Meza Lasso et al., 2016; Carbonell et al., 2018), the common white mouse (Salazar and Arango, 2014) and zebrafish (Akle et al., 2017). Even though several amphibians are frequently used as vertebrate models to study development and regeneration, only a few studies report the use of amphibians such as Xenopus laevis) (Daudin, 1802) in electrophysiological studies (Chaves et al., 2003). In the field of regenerative biology, amphibians represent one of the key groups of animals in which to study tissue regeneration. Although representative studies in this area have been carried out in our country, there is a clear need to continue expanding our knowledge in this discipline (Arenas et al., 2015). Therefore, the implementation of animal models for research in developmental and regenerative biology is a task that must be carried out.
The established salamander models to study regeneration are Ambystoma mexicanum (Shaw and Nodder, 1798), Notophthalmus viridescens (Rafinesque, 1820), Cynops phyrrogaster (Boie, 1826), and Pleurodeles waltl (Michahelles, 1830) (Joven et al., 2019), with A. mexicanum being the most commonly used model because its generation time is shorter than other salamanders (10 - 18 months) versus N. viridescens (3 years) (Farkas and Monaghan, 2015; Simon and Odelberg, 2015). and they reproduce easily in laboratory conditions. A. mexicanum has a broad capacity to regenerate different tissues such as heart, lungs, elements of the central and peripheral nervous system, gut, jaws, tail and limbs (Brockes and Kumar, 2008). In addition to the ability to regenerate multiple structures, this species represents a tool for the study of developmental biology given its short time to sexual maturation and relatively short time to embryonic development, among other virtues (Table 1). Currently, different tools such as CRISPR/Cas9, plasmid electroporation, and morpholinos, as well as the creation of transgenic lines have been applied in this model to understand the molecular and cellular inputs during tissue regeneration (Echeverri and Tanaka, 2003; Sobkow et al., 2006; Voss et al., 2009; Khattak et al., 2013, Fei et al., 2014, 2018;) (Table 2). Also, one of the major advances in the field of study of axolotls was the work on the sequencing and assembly of their genome at the chromosomal level (Nowoshilow et al., 2018a; Smith et al., 2019). The assembly of the axolotl genome will facilitate the analysis ofgene structure, function, and the intricate epigenetic layers underlying transcriptional regulation, fundamental for our understanding of vertebrate evolution, development, and tissue regeneration. Considering all these characteristics, Ambystoma mexicanum is an excellent model to answer different questions that we can raise from developmental biology and regeneration (Table 1). The advances in this field could impact other disciplines such as developmental malformations, teratogenesis, tissue engineering, and regenerative medicine. Likewise, it benefits the creation of academic scenarios for the training of professionals in biology and other related areas in biomedical sciences, giving the opportunity for collaborative studies at the national and international level.
Table 1 Main disciplines where the A. mexicanum can be used to answer biológica! research problems
| Areas of study | Features of the model | Research question | References |
|---|---|---|---|
| Limb development | Salamanders undergo preaxial domi-nance during the development of the skeletal elements | Which are the molecular mechanis-ms that trigger the morphogenetic changes during limb development through evolution in salamanders? | (Frõbisch et al., 2015; Frõbisch and Shubin, 2011; Kerney et al., 2018) |
| Paedomorphism | Thyroid hormone mediated the plastici-ty during development in salamanders | Which are the molecular mechanis-ms that regulate neoteny in axolotls? | (Crowner et al., 2019; Huggins et al., 2012; Johnson and Voss, 2013; S. R. Voss et al., 2012) |
| Cellular senescence | Crosstalk between the senescent cells and immune system as a crucial input for limb regeneration | How senescent cells recruit the im-mune cells to the wound to favor tissue regeneration? | (Davaapil et al., 2017; Yun, 2018; Yun et al., 2015) |
| Limb regeneration | Salamanders are the only tetrapod with the capacity to regenerate limbs throu-ghout their lives. | Which are the cellular and molecular mechanisms that allow limb regene-ration in salamanders? | (Bryant et al., 2017; Tobias Gerber et al., 2018; Haas and Whited, 2017; Kragl et al., 2009; S. Randal Voss et al., 2015) |
| Regeneration of Neural Tissue | Regenerate complex networks of neural tissues such as injuries in the spinal cord and brain using a pool of stem cells. Cellular plasticity to regenera-te lens and retina through cellular transdifferentiation. | How is the wound microenvironment modulated to induce a regenerative response in the spinal cord injury? Which is the molecular signature that allows a transdifferentiation process during lens regeneration? | (Amamoto et al., 2016; Freitas et al., 2019; Joven and Simon, 2018; Sabin et al., 2019; Suetsugu-Maki et al., 2012; Tazaki et al., 2017; Tsonis and Del Rio-Tsonis, 2004) |
| Heart regeneration | Model of myocardial infactation which allow to understand the remodeling of ECM and cardiomyocytes proliferation during heart damage | How the immune system allows a permissive microenvironment for cardiomyocytes proliferation? | (Cano-martínez et al., 2010; Godwin et al., 2017) |
| Oral-facial regeneration | Hight capacity to regenerate soft tissue like cartilage. | How oral tissue regeneration can be achieved in axolotls? | (Charbonneau et al., 2016) |
| Cancer | The cellular and molecular mechanisms during tissue regeneration in salamanders are similar to the mechanisms that allow cell transformation in a tumor. | Why the cells present during a regenerative process are more resistant to form tumors? | (Brockes, 1998; Fior, 2014) |
Table 2 Experimental biology techniques that support the versatility of the Ambystoma mexicanum model
| Biological tools | Application | References |
|---|---|---|
| Trangenesis | A. mexicanum is a suitable model to do Knock in or Knock out animals to test the function of genes. The use of different tools has been used such as I-SceI meganuclease, Tol2 transposase, LacI-Inducible System CreER/LoxP and CRISPR-Cas9 | (Fei et al., 2014, 2018; Khattak et al., 2014; Khattak and Tanaka, 2015; Sanor et al., 2020; Whited, 2012) |
| Chemical screening | Different chemical can be used to inhibit important signal pathways during development and regeneration in axolotl such as Wnt, Nocht, FGF. | (Baddar et al., 2021; Larissa V. Po-nomarevaa, Antony Athippozhyb, Jon S. Thorsona, 2015) |
This review is focused on A. mexicanum, commonly named axolotl. The general biological features of this model organism are presented; also, the description of the general protocol used to establish a functionally breeding colony in the laboratory and perspectives on use of this model in basic biology research in Colombia to understand the molecular and cellular mechanisms driving development and tissue regeneration are presented.
THE MEXICAN AXOLOTL: A MODEL ORGANISM WITH A LONG HISTORY
Salamanders (tailed amphibians) represent one of the three major amphibian groups, the other two being caecilians and anurans (frogs and toads). These groups have been evolving independently for more than 200 million years and are highly divergent from one another (Wiens, 2007). The A. mexicanum belongs to the family Ambystomatidae, which is comprised of 32 species distributed from Mexico to Canada; particularly, A. mexicanum is closely related to the tiger salamander (A. tigrinum) according to the phylogeny of the Ambystomatid salamanders proposed by Shaffer (1993). Axolotls are thought to have originated from metamorphic tiger salamander ancestors that colonized large lake systems once typifying the natural habitat ofpresent-day Mexico City (Brandon, 1989; Shaffer, 1993). Axolotls are characterized by being paedomorphic; they naturally do not undergo metamorphosis, thus retaining larval characteristics even when they have reached sexual maturity (Brandon, 1989).
They are endemic to Lake Xochimilco in the Mexico Central Valley, this high-altitude lake system (~2200 m. a. s. l.) has been inhabited for centuries, most notably by the Aztecs (Contreras et al., 2009). The lake has been reduced over the centuries, and is currently a 40 km2 area of artificial canals just outside the city limits of Mexico City; therefore, A. mexicanum has suffered a dramatic population reduction due to habitat transformation and is currently classified as a critically endangered species with an ever decreasing trend for wild populations (Contreras et al., 2009). Nowadays, the largest colonies of A. mexicanum are in different laboratories around the world.
One of the earliest mentions of A. mexicanum in research literature is from 1865, when this species was exhibited for the first time in the Natural History Museum in Paris, France. From this time, publications reporting the use of A. mexicanum began to increase, and between 1920 and 1960 the maximum scientific production occurred in areas such as embryology, regeneration, metamorphosis, endocrinology, radiation, transplantation, evolution, and anatomy (Shaffer, 1993; Keinath et al., 2015). The axolotl provides a tractable system for embryological manipulations and the study of pattern formation (Shaffer, 1993; Nacu et al., 2013; Fei et al., 2014, 2018), due to the size of the embryos and its cells it is relatively easy to observe early cleavage of cells and morphogenesis under a microscope. Several of its advantages lie in the easy reproduction, obtaining numerous offspring, straightforward husbandry in the laboratory and the size of their eggs allows better handling favoring the use of microinjection techniques for the creation of transgenic lines (Sobkow et al., 2006; Khattak et al., 2009). Also, the axolotl can regenerate different tissues, organs and structures throughout its lifespan, which places it as an ideal vertebrate model for the study of regenerative biology (Roy and Gatien, 2008; McCusker and Gardiner, 2011), in order to understand the cellular and molecular mechanisms that underlie tissue regeneration (Table 1).
Among vertebrates, axolotls are one of the organisms with the largest genome (-32 GB), ten times larger than the human genome. Thanks to the efforts of different laboratories around the world, the reference genome was published in 2018 (Nowoshilow et al., 2018; Smith et al., 2019). One of the main reasons of behind the large genome size in axolotls is the presence of repetitive sequence like the long terminal repeat (LTR) retroelement (Keinath et al., 2015) which represent around 18 GB of the 32 GB genome size of the axolotl (Nowoshilow et al., 2018). Interestingly, a lot of these repetitive elements are not located in introns that make part of important developmental genes, which probably is under evolutionary constraint in axolotls to allow the rapid transcription of important genes for development (Nowoshilow et al., 2018).
While it is true that the large size of the genome in this species may represent an apparent barrier to genetic analysis, it is also important to note that its knowledge will improve the efficiency of various techniques such as CRISPR-mediated gene editing, TALENS, and other editing systems. On the other hand, the knowledge of genomic sequences creates the opportunity to decipher the epigenetic mechanisms involved in development and regeneration, as well as the protein-DNA interaction and consequently the regulation of gene transcription.
HUSBANDRY AND REPRODUCTION: THE COLOMBIAN EXPERIENCE
In Colombia the field of developmental biology and regeneration is a nascent field with few laboratories currently dedicated to the study of animal and plant development (Marcellini et al., 2017). In the laboratory of Genetics, Regeneration and Cancer (GRC) at Universidad de Antioquia, the first colony of A. mexicanum in Colombia has been established with the first animals coming from the Ambystoma Genetic Stock Center (AGSC) at the University of Kentucky in United States of America (Smith et al., 2005). Colombia has a broad environmental legislation regarding genetic resources and introduction of foreign species for research. In order to establish the colony, a process of sending permits for use of genetic resources and importation of the animals required to establish the colony was done to the national environmental authority, ANLA.
Establishment of the colony
The colony of A. mexicanum was established in the laboratory, following different protocols already available (Kim, 1996; Farkas and Monaghan, 2015). A total of 30 animals were imported from the AGSC approximately six years ago. Animals were imported under the CITES # 14US436S065B/9. The total of 30 animals comprised ten albinos, ten leucistic, and ten wild type of size 6-8 cm. Each animal was housed in plastic containers (19 cm x 14 cm x10 cm) and were feed every day with salmon pellets (FLYHOUSE, NY USA) in order to increase the growth rate using a light/ dark period 12/12. Currently, our laboratory has around 300 juvenile animals and more than 100 growing larvae of which 100 juveniles have been used in experimental procedures within approved research projects. 70 % of these animals are wild type, while the remaining 30 % comprise 20 % leucistic and 10 % with albino phenotype. Several of the phenotypes currently available in our laboratory are shown in Fig. 1a-1f.
Maintenance of adult axolotls
The adults are kept separately in plastic tanks (53 cm x 30 cm x 23 cm) with 15 L of water at 19 - 21 °C. Tap water is used, which is filtered with a MILIPORE purification blue canister system (5 jm, 1 jm and activated carbon filters) to remove heavy metals and chlorine. The water is changed twice a week after the animals are feed; if the water is cloudy at the time of feeding, then it is changed prior to feeding. Adults are fed with Sinking Feeder Pellets. To avoid the proliferation of microorganisms, the tanks should be disinfected with Virkon every two weeks. If the water is very cloudy due to animal waste, it should be changed because ammonia accumulation could be toxic to the animals.
Reproduction ex situ
Fertilization in axolotls is internal; to promote natural reproduction, a male and a female are placed together overnight at 13 - 16 °C by placing cold gel bags in the plastic tank (Aguilar-Miguel et al., 2009). This colder temperature is thought to promote reproductive behavior. After a courtship ritual, in which the male places himself on the female's dorsal area to stimulate her, the female incorporates in her cloaca a spermatophore left by the male on the bottom tank.
Plastic plants are placed in the water, so the female can have a place to lay the eggs (Khattak et al., 2014), given that it has been reported that even though most Ambystomatids leave their eggs without any parental care (Bruce, 2003), axolotls need a plant-like structure for egg laying (Zambrano et al., 2007). Artificial leaves and branches are placed in the tank, also to give a more "naturalized" environment, in which the female can lay the eggs. The female takes around 24 hours to lay all the eggs, usually between 100 and 400 eggs (Gresens, 2004; Eisthen and Krause, 2012). In our experience the best season to get good quality fertilized eggs is between February and May, between August and October few fertilized eggs will be produced. The PMX population management software (Frankham et al., 2017) was used to make very careful breeding choices according to mean kinship and maximal avoidance of inbreeding, this analysis was performed by AGSC, University of Kentucky (USA).
Breeding ex situ
After fertilization, the eggs are taken from the plastic leaves and put in plastic containers with Holtfreter's salt solution (20 %). The larvae breeding consists of feeding the larvae with brine shrimp (Artemia sp.) when the yolk in their bellies disappear. They are fed daily until they reach a size of around 4 cm (Khattak et al., 2014), then we start to feed them with crushed fish pellets three times per week. The brine shrimp eggs hatch in salt solution media at a temperature of 28-32 °C, and a water aeration system is also used (Browne et al., 2000). The hatched shrimp are put ad libitum in the container where the larvae are kept and the Holtfreter's salt solution is changed the next day.
Cannibalism in larvae of some species of Ambistomatidae has been reported; for A. tigrinum (Lannoo et al., 1989), A. macrodactylum (Wildy and Lynn, 2000), and A. mexicanum cannibal behavior has been observed mainly in the larval and juvenile stage, even if the larvae have an adequate food supply and are maintained at a recommended density (Robles-Mendoza et al., 2009). Cannibalism may increase if A. mexicanum larvae are more concentrated in a small space, even if the food sources are sufficient. Once the larvae have developed completely, they are fed with "fish food pellets", as A. mexicanum displays distinct changes in its feeding behavior with increasing larval age (DeNardo, 1995; O'Rourke, 2002).
Axolotl larvae are maintained in Holtfreter's salt solution. Robles-Mendoza et al. (2009) recommend the use of Holtfreter's solution for the maintenance of early stages of A. mexicanum on culture systems, because this solution meets better their physiological requirements, as the highest growth was registered with Holtfreter's salt solution, compared to other maintenance media for axolotl juveniles, such as dechlorinated tap water and dechlorinated tap water enriched with sodium chloride and commercial colloidal solution.
Development
Whereas many tiger salamanders undergo an obligatory metamorphosis, axolotls rarely, if ever, transform in a laboratory. Therefore, axolotls retain larval features into adulthood and therefore require aquatic husbandry procedures throughout life. However, it is possible to induce metamorphosis in axolotls by adding thyroid hormone to the rearing water (Page and Voss, 2009).
In the usual development of A. mexicanum, 40 distinctive stages have been described, including cleavage stages, blastula stages, gastrula stages, neurula stages, and late embryo development stages, which end with the egg hatching (Schreckenberg and Jacobson, 1975). Through cleavage (Fig. 2a-2d), blastula (Fig. 2e-2f), and gastrula stages (not displayed here), the embryos rest with their animal poles uppermost, then at the end of stage 11, the embryo rotates turning the future dorsal side uppermost, remaining in this orientation from stage 12 to 19 (Schreckenberg and Jacobson, 1975). As neural tube formation (Fig. 3a-3h) approaches completion at stage 19, the embryo begins to lie on its side. It is completely on its side from stage 20 to stage 39 (Fig. 4a-4h). Finally, at stage 40 (Fig. 4i), which is reached after several days, larvae have completely turned onto their bellies, and at this point they hatch (Schreckenberg and Jacobson, 1975).

Figure 2 Stages of initial embryonic developoment (blastula formation) in Ambystoma mexicanum. a). Egg, b). Fertilized egg (stage 1), c). First cleavage, 2 cells (stage 2), d). 4 cells (stage 3), e). 8 cells (stage 4), f). 32 cells (stage 6), g). 64 cells (stage 7), h). Early blastula (stage 8).
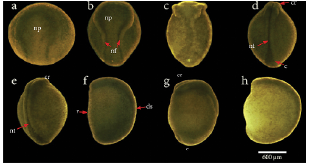
Figure 3 Neural tube development stages in Ambystoma mexicanum. a). Early neurula (stage 14), b). Middle neurula (stage 16), c). Late neurula I (stage 17), d). Late neurula II (stage 18), e). Late neurula III (stage 19), f). Late neurula I, lateral view (stage 17), g). Late neurula II, lateral view (stage 18), h). Late neurula III, lateral view (stage 19). s c, caudal; cr, cranial region; ds, dorsal; np, neural plate;neural folds; nt, neural tube ; v, ventral.
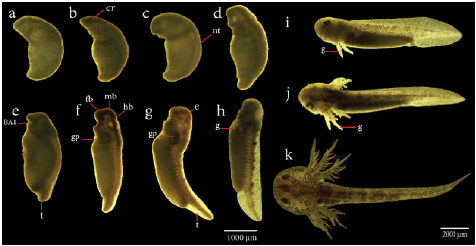
Figure 4 Late embryonic development and organogenesis of Ambystoma mexicanum. a). Stage 23, b). Stage 25, c). Stage 27, d). Stage 29, e). Stage 31, f). Stage 32, g). Stage 36, h). Stage 39.(i-k), Sequence of photographs of the last developmental stages including gill formation of A. mexicanum, taken from wild type embryos. i). Stage 40, j). Stage 43, k). Stage 44. BA1, first branchial arch; c, caudal; cr, cranial region;e, eyes ;fb, forebrain; g, gills; gp, primordium gills; hb, hindbrain; mb,midbrain; t,tail.
Limb and tail regeneration
As axolotls have the feature to regenerate perfectly lost limbs and other structures, some authors have suggested that mechanisms involved in limb regeneration are similar to those controlling larval limb development (Carlson et al., 2001; Ghosh et al., 2008). A staging system for the forelimb regeneration in the axolotl has been established. It begins with the wound healing stage which concludes at 3 days post amputation (dpa) followed by dedifferentiation (8 dpa), early bud (15 dpa) (Fig.5a), medium bud (17 dpa) and late bud formation (20 dpa) (Fig 5b). The process concludes with palette formation (28 dpa) (Fig 5c) and digital outgrowth (32 dpa) (Fig. 5d-5e) (Tank et al., 1976). However, this timing varies depending on the influence of different factors such as age and size of the animal (Seifert et al., 2012; Monaghan et al., 2014). The staging system of forelimb regeneration can be compared in anatomical terms to the stages of limb development in A. mexicanum which have also been described (Nye et al., 2003).
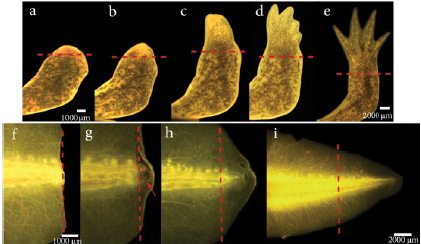
Figure 5 Limb and tail regeneration in Ambystoma mexicanum. (a-e) Limb regeneration a). Early blastema b). Late blastema c). Palette stage d). Digital outgrowth e). Completed regeneration. (f-i) Tail regeneration. f). Wound healing, 24 hours post-amputation. g). Blastema development, the red arrow indicates the blastema. h). Tail outgrowth 14 dpa. i). Completed regeneration. The dotted line represents the amputation plane.
The axolotl has an extraordinary capacity to regenerate other appendages such as the tail (Clarke et al., 1988; Echeverri et al., 2001; Echeverri and Tanaka, 2002; Mchedlishvili et al., 2012; Ponomareva et al., 2015). The tail of these animals is made up ofvarious tissues such as muscle, cartilage, connective tissue (dermis), and the spinal cord (Echeverri et al., 2001). The study of the tail regeneration capacity in A. mexicanum has gained great relevance because the spinal cord regenerates, which creates an opportunity to study cellular and molecular mechanisms that regulate this event (Chernoff et al., 2003); allowing the design of new regenerative therapies to regenerate spinal cord injuries (Thygesen et al., 2019).
The regeneration of tail and limbs in axolotls comprises three stages: wound healing, development of the blastema, and ends with morphogenesis and growth (Echeverri et al., 2001; Simon and Tanaka, 2013). In the laboratory we are currently working with animals between 8-10 cm (~ 9 months of age), where wound healing has been observed 24 hours after amputation, blastema formation is evident at 7 dpa (early blastema) until 9 dpa (late blastema). The growth and morphogenesis phase take place between 11 dpa and 42 dpa when the total size of the amputated tissue is regenerated (Fig. 5f-5i). However, the time in which these stages occur differs from those of smaller animals with approximately 2-3 cm (Echeverri et al., 2001). Currently, in our laboratory, several ongoing projects are evaluating the effect of some signaling molecules involved in the regeneration of this structure such as reactive oxygen species (ROS) and their interaction with other signaling pathways, using chemical inhibitors that are placed directly on the aquatic environment (Carbonell et al., 2021). Given the location of the tail, its shape, and the relatively easy surgical access to the tissue, it allows tools such as electroporation of plasmids and other constructs to be quite viable (Echeverri and Tanaka, 2003). This highlights the axolotl tail regeneration model as a powerful tool to study the different events that underlie this regenerative process.
Perspectives for Ambystoma mexicanum as a model organism in Colombia
Colombia is considered a megadiverse country with 806 reported amphibian species; amongst them 25 species of salamanders are reported (Acosta-Galvis and Gutiérrez-Lamus, 2012) and all of them belong to Plethodontidae, or lungless terrestrial salamanders, with two representative genera, Bolitoglossa with 23 species and Oedipina with two species in Colombia (Galeano et al., 2006; Acosta-Galvis and Cuentas 2019). Efforts to study Colombian plethodontids salamanders in different fields as evolution and phylogenetics have been reported (Parra-Olea et al., 2004; Silva et al., 2011). Also, morphological, histological and molecular aspects of limb regeneration in the wild species Bolitoglossa ramosi have been described (Arenas Gómez et al., 2017; Arenas Gómez et al., 2018), aiming to compare it with traditional model salamanders of the families Ambystomidae and Salamandridae and eventually, to establish B. ramosi as a new model organism for regeneration, to understand how conserved limb regeneration is between salamander families. Husbandry of terrestrial plethodontid salamanders in a laboratory environment have been attempted with B. ramosi and Bolitoglossa vallecula in an attempt to use them as models in regeneration (Arenas Gómez et al., 2015); however, these species require specific conditions for reproduction which have to be elucidated. Currently, the difficulty to reproduce these species in a laboratory environment constitutes the main barrier to use them as model organisms in regeneration. Also, Colombian legislation to carry out research with biodiversity requires a complicated process for genetic resources access.
Why use A. mexicanum, as a model given the number of amphibians with unknown features found in Colombia? The answer is complex, not all amphibians have the capacity to regenerate, and the best "regenerators" amongst amphibians, are the salamanders. In this context, A. mexicanum, is an excellent model organism as it is easy to handle and reproduces in the laboratory (Roy and Gatien, 2008) and the research done on this species is considerable (Voss et al. 2009; McCusker and Gardiner 2011; Dwaraka and Voss, 2019; Joven et al., 2019) such as the insight in the behavior of cells during controlled growth and pattern formation leading to regeneration (Kumar et al., 2007; Roy and Gatien, 2008; McCusker and Gardiner, 2011; Simon and Tanaka, 2013; Tanaka, 2016). Moreover, axolotls have shown that their regenerative potential endure throughout their lifespan. In addition, this animal model presents important characteristics such as low risk to develop cancer and senescence resistance, which makes the axolotl an ideal organism to understand biological processes such as aging and cancer (Brockes, 1998; Khudoley and Eliseiv, 1979; Vieira et al., 2019).
On the other hand, considering the growing interest in our country to study the different mechanisms involved in developmental malformations, and the desire to strengthen studies in developmental biology and regeneration for the design of new therapeutics in regenerative medicine, the Ambystoma mexicanum model meets all the characteristics to respond to these needs (See table 1, table 2 and table 3). Throughout history, Colombia is a country that has been characterized by predominantly conducting studies focused on areas such as Ecology, Biodiversity of fauna and flora, and taxonomy is given our great wealth of natural resources. However, being coherent with the new challenges of science and without detracting from the aforementioned works, it is pertinent that in order to be more competitive at an international level in fields such as regenerative medicine and biotechnology, studies in the field of experimental biology, including developmental biology and tissue regeneration, should be strengthened and increased. The above appreciation stems from the fact that several animal models and in particular regenerative models, provide knowledge that can promote the development of new therapies in the field of regenerative medicine and other disciplines. Our experience has shown that after the implementation of the Ambystoma animal model, national and international collaborations have increased with different research groups in various areas of knowledge such as Medicine, Biology, Dentistry, Biotechnology, Neurosciences, and Materials Engineering from student internships to collaborative projects.
In the Genetics, Regeneration and Cancer research group at Universidad de Antioquia, a project with A. mexicanum is currently being developed; the project includes the standardization of conditions to develop transgenic lines (e.g., GFP transgenic animals) which could be the first line of transgenic vertebrates generated in Colombia. Additionally, the research group is studying the cellular and molecular mechanisms that are involved in the in vivo regeneration of complex structures such as limbs and tail regeneration.
Table 3 Comparison between Ambystoma mexicanum model with other main developmental models
| Animal model | Embryonic period / Embryonic manipulation | Sexual maturation time | Sequenced Genome | Adult Regenerative Capacity | Loss and gain of gene function assays/ Development of transgenic animals | Chemical screen during development | References |
|---|---|---|---|---|---|---|---|
| Ambystoma mexicanum | 18 - 20 days with high quantities of embryos / Direct visualization of embryonic development by stereomicroscopy and tissue transplantation can be performed | 8-12 months | Yes | Yes / Heart, Brain, Spinal cord, Lungs, lens, tails, Jaws and Limbs | Different tools for the generation of transgenic animals can be used as I-SceI meganuclease, Tol2 transposase, LacI-Inducible System CreER/LoxP and CRISPR-Cas9. | Facility to evaluate teratogenic agents and chemical inhibitors during development | (Nowoshilow et al., 2018b; Ponomareva et al., 2015; Smith et al., 2019; Voss et al., 2009) |
| Gallus gallus | 21 days / Easy embryo manipulation and tissue transplantation | 6-8 months | Yes | Limited to Hair Cels | (RCAS)system, knockdown with siRNA (Small interference RNA) and Morpholinos / Tol2 System, TALENS or CRISPR/Cas9 | Different toxicity and teratogene-sis tests can be performed on the developing embryo | (Sánchez Alvarado and Tsonis, 2006; Bellairs and Osmond, 2014; Jelinek, 1982; Stoller and Fekete, 2016) |
| Danio rerio (Zebra fish) | 48 - 72 hours / direct visualization of embryonic development due to its high transparency | 2-3 months | Yes | Yes/ Fins, tail, heart, liver, spinal cord, hair cells of inner ear and Jaws | Morpholinos, siRNA, Chemical inhibitors / Tol2 System, Mutage-nesis, Cre-LoxP system, TALENS or CRISPR/ Cas9 | Various toxicity, teratogenesis and signaling pathway in-hibitor assays are widely performed. | (Hogan et al., 2008; Kimmel et al., 1995; Lin et al., 2013; Marques et al., 2019) |
| Xenopus | 15- 16 days (Premetamorphosis) / easy visualization of em-bryonic development and tissue transplants | 7-8 months | Yes | Yes/ In pre-me-tamorphic state: limbs and tail. In adult hair cells of the inner ear | Interference RNA, Chemical inhibitors/ Tol2 System, Cre-Loxp, TALENS or CRISPR/ Cas9 | Teratogenic substances and signaling pathway inhibitors can be used | (Blum and Ott, 2018; Heasman, 2006; Horb et al., 2019; Lavery and Hoppler, 2008; Phipps et al., 2020) |
CONCLUSIONS
In Colombia, regenerative biology is an emerging research field and the Mexican axolotl, A. mexicanum, has proven to be a suitable model to understand several biological processes, including regeneration. Their husbandry is simple, inexpensive, and has high and frequent offspring production. Also, the release of the genome of A. mexicanum, represents an important advance in the field of regeneration because this would allow to use this organism in different functional assays to understand molecular and cellular mechanisms which allow tissue regeneration in salamanders and to develop, in the future, cell therapies that can help in the treatment of different diseases in humans.
















