INTRODUCTION
The World Health Organization report that, in 2016, children under the age of five mainly died of diarrhea, acute respiratory infections, and malaria 1. These types of diseases represent a greater burden for lower-income countries, due to flaws in food sanitation, hygiene, and safety, which facilitates pathogen propagation, especially enteric ones 2. Among infectious diseases, those of bacterial etiology are being associated with environmental disinfection and its health conditions, more specifically, surface contamination facilitates the propagation of pathogens among children and family members 3,4.
It is known that the health impact of environmental microbiological exposure is greater in children under the age of five, due to physiological immaturity, and deficient hygiene habits, such as hand washing and adequate toilet use. Similarly, child care centers contain greater indoor concentration (3870 UFC/mm3) of bacteria, when compared to schools and older adult care centers 5 and how to effectively prevent these, remains. his study aimed (i. he air in these environments presents a wide range of bacteria, such as Bacteroidetes, Bacillus, Ruminococcus, Coprococcus, Ente-robacter, Flavobacterium, Staphylococcus, Micrococcus, and Corynebacterium6.
One study identified that Bacillus, Staphylococcus, Brevibacillus, Pseudomona, Moraxella, Enterococcus, Acinetobacter, and Microbacterium isolate on the toys and furniture of day care centers 7. Furthermore, Pseudomonas, Pantoea, Bacillus, Staphylococcus, and Streptococcus have been identified in home refrigerators and bathrooms 8) whereas in classrooms, Sphingomonas, Staphylococcus, and Enterobacteria prevailed 9.
Even though most of these bacteria are not primary pathogens, some can survive and be transmitted through the surfaces of indoor environments, and cause community-acquired infections. hese bacteria may be nosocomial, and present multidrug resistance, as is the case of methicillin-resistant Staphylococcus aureus (MRSA) 10. However, genetic studies with community-acquired MRSA strains have revealed differences in relation to those of nosocomial origins, demonstrating that they are susceptible to many other antibiotics, such as tobramycin, gentamycin, lincomycin, cloranfenicol, vancomycin 11. Studies have shown an increase of community-acquired MRSA among children. hese strains are not multi-resistant, but can present additional resistance to erythromycin 12. Other community-acquired pathogens that are multi-resistant to antimicrobials are E.coli and Klebsiella spp. These are common causing agents of urinary infections, with a high percentage of extended-spectrum beta-lactamase producing bacteria 13Rio Grande do Norte State capital, northeastern Brazil, from 2007 to 2010. A total of 1,082 positive samples were evaluated; E. coli was the most prevalent pathogen (60.4%. Possible factors associated with the prevalence of antimicrobial resistance in the community are the inappropriate prescription of antibiotics, the genetic variability of bacteria, and the genetic expression of inducible resistance; situations that require further study 10.
Considering the influence of the environment in transmitting infectious diseases among children, the problem of bacterial resistance, and its relevance to public health, the objective of this study was to characterize the pattern of antibiotic resistance of the bacterial biota isolated from the surfaces of child care centers.
MATERIALS AND METHODS
Study design and area: This was a cross-sectional descriptive study, conducted in 2016, in 230 government-funded child care centers located in the urban perimeter of Bogotá, Colombia. The city had 897 public child care centers, and a population of 11.973 children, between the ages of 2 and 5, at the time of data collection. Sample size was calculated using Win Episcope 2.0® software (University of Edinburgh, 2000), set at 266 child care centers by assuming a 50% prevalence of infectious agents, confidence level of 95%, and an expected margin of error of 5%. Probabilistic sampling was used, and informed consent was given by all 266 centers, ensuring voluntary participation. This study was approved by the Research Ethics Committee of the Universidad Nacional de Colombia, and was developed according to the principles enshrined in the Declaration of Helsinki.
Sample collection: First, the researchers verified that the facilities contained the following items: at least one bathroom, one kitchen, and a play and eating area for the children. Samples were taken in the first hours of the morning, before the arrival of the children, and then, following the cleaning and disinfection protocol indicated by the person in charge of each center. Samples were taken from surfaces in the kitchens, bathrooms, and places to which children were directly exposed, spent most of their time in, and carried out their play activities (chairs, tables, mattresses, toys, and colored pencils). Swab samples were collecting using the COPAN Venturi Transystem® (Innovation, Italy) (STUART sterile transport swab system), which were then transported and processed up to eight hours after collection in the Laboratory of Integrative Health of the Universidad de La Salle, where all microbiological analyses took place. The personnel in charge of sample collection were previously trained, and they wore all the necessary personal protective equipment to avoid cross-contamination (disposable caps, gloves, mouth masks, and aprons). The protocols were conducted according to the Standard Operation Procedures (SOP) validated in the lab in previous studies.
Antimicrobial susceptibility profile: The samples were seeded in blood and MacConkey agar, and incubated at 37° C for 24 hours. Bacterial colonies were classified as either Gram-positive or Gram-negative, using Gram staining, and were isolated in trypticase soy agar, under the same incubation conditions. Microbiological typing of the isolates and antibiotic susceptibility was carried out using the Vitek® 2 compact computerized system (Biomerieux, Marcy l'Etoile, France) with Vitek 2-GP cards® (Reference 22218 for Gram-positive), Vitek® 2- GN cards (Reference 414163 for Gram negative), Vitek® 2 AST-P577 (Gram-positive susceptibility), and Vitek® 2 AST-NO82 (Gram-negative susceptibility), according to the protocol described by Herold et al. 1998 14 To analyze antimicrobial susceptibility, the software used cutoff points established in the Clinical and Laboratory Standards Institute 15. Moreover, as recommended by the manufacturer for antimicrobial identification and susceptibility, the following ATCC strains were used for quality control: Klebsiella oxytoca ATCC® 700324, and Staphylococcus aureus ATCC®29213. The results were analyzed using descriptive statistics, calculated in terms of frequencies and percentages.
RESULTS
The pattern of antimicrobial susceptibility of the 151 bacteria, isolated from the environments of 266 childcare centers, was analyzed (72 Gram-positive and 79 Gram-negative bacteria). The greatest number of bacteria were found on kitchen surfaces (n=58), followed by bathrooms (n=38), classrooms (n=35) and mattresses, toys, and colored pencils (n=20).
The most common genus of bacteria was Staphylococcus spp (72 isolates). Furthermore, 94.4% of these isolates (68/72) were coagulase-negative Staphylococcus (CNS), and the most representative species was S. epidermidis, with 23 isolates, followed by S. haemolyticus (14/72). Additionally, 5.6% were identified as S. aureus (4/72). Table 1 shows the distribution of these bacteria among the studied surfaces. [Table 1].
Table 1 Frequency of isolates by surface analyzed in the early childhood day care centers
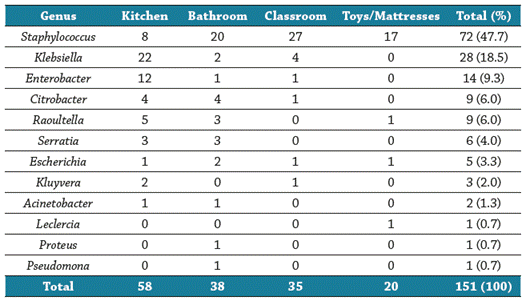
Source: elaboration of the authors.
The most common Gram-negative bacilli were Klebsiella (K. oxytoca and K. pneumoniae), Entero-bacter (E. cloacae, E. aerogenes, E. amnigenus and E. asburiae), Citrobacter (C. freundii, C. koseri and C. amalonaticuss), and Raoultella (R. ornithinolytica and R. planticolaa) (Table 1).
Regarding the analysis of antimicrobial resistance patterns, species belonging to the Staphylococ-cus genus were resistant to some of the analyzed antibiotics, except for S. simulans. he highest percentages of resistance were found against erythromycin (48.6%), oxacillin and tetracycline (26.4%), cephoxytine (23.6%), and clindamycin (20.8%). None of the strains were quinolone-resistant (Table 2). Two S. hominis strains (2.7%) presented Methicillin-resistant phenotypes (MRS), which originated from bathrooms and toys/mattresses. Seventeen strains of Staphylo-coccus (23.6%), isolated, primarily, from toys/mattresses and classrooms, presented multidrug resistance, especially to beta-lactamases, macrolides, and clindamycin (Table 3).
Table 2 Number of antibiotic-resistant strains of Staphylococcus, isolated from surfaces in early childhood day care centers
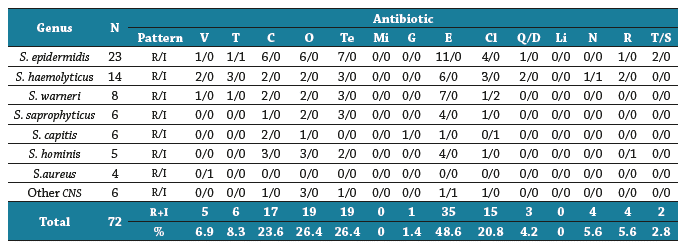
R/I: Resistant/Intermediate. V: Vancomycin, T: Teicoplanin, C: Cefoxitin, O: Oxacillin, Te: Tetracycline, Mi: Minocycline, G:Gentamicin, E: Erythromycin, Cl: Clindamycin, Q/D: Quinupristin/Dalfoprisitn, Li: Linezolid, N:Nitrofuran-toin, , R: Rifampicin, T/S: Trimethoprim/Sulfamethoxazole.
Source: elaboration of the authors.
Table 3 Patterns of resistance of Staphylococcus strains isolated in this study
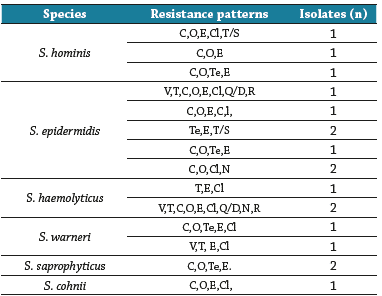
V: Vancomycin, T: Teicoplanin, C: Cefoxitin, O: Oxacillin, Te: Tetracycline, E: Erythromycin, Cl: Clindamycin, Q/D: Quinuspristin/Dalfoprisitn, N:Nitrofu-rantoin, R: Rifampicin, T/S: Trimethoprim/Sulfamethoxazole.
Source: elaboration of the authors.
The Gram-negative bacilli identified in the study were most resistant to ampicillin (57.0%) and cephalosporins (cephalothin - 49.4%, Cefuroxime Axetil - 34.2% and Cephuroxime - 30.4%). Enterobactereacea isolates did not display multidrug resistance; however, most were resistant to cephalosporin. Of the non-fermenting Gram-negative bacilli, Acinetobacter was resistance to be-ta-lactamases and nitrofurantoin. he only isolate of P. aureginosa in this study was multidrug resistant (beta-lactamases, nitrofurantoin, and Trimethoprim/Sulfamethoxazole) (Table 4).
Table 4 Number of antimicrobial resistant strains of Gram-negative bacilli isolated from surfaces in early childhood day care centers
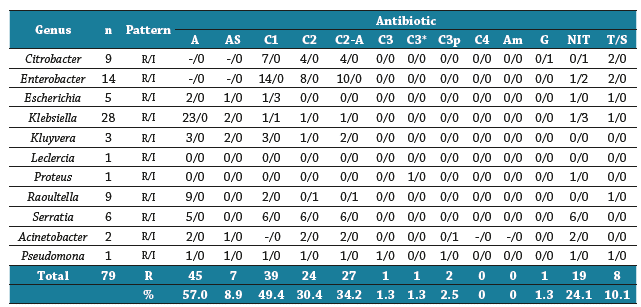
R/I: Resistant/Intermediate. A: Ampicillin, AS: Ampicillin/sulbactam, C1: Cephalothin, C2: Cefuroxime, C2-A: Cefuroxime Axetil, C3: Cefotaxime, C3*: Ceftazidime, C3p: Ceftriaxone, C4: Cefepime, Am: Amikacin, G: Gentamicin, Nitrofurantoin, T/S: Trimethoprim/Sulfamethoxazole. In the table: (-) not assessed.
Source: elaboration of the authors.
DISCUSSION
Child care centers can represent transmission sites for infectious diseases, especially because of the exposure of children to surfaces considered reservoirs for microorganisms. In the present study, the most common isolates found in classrooms, bathrooms, mattresses, and toys was Staphylococcus spp, especially CNS. his finding is consistent with that of previous studies, which have shown that these environments contain high levels and diversity of microorganisms, with a predominance of phyla Firmicutes and Proteobacteria, and genera Bacillus and Staphylococcus6,7.
CNS have been isolated from the hands of children 16, and more generally, from skin and mucous membrane microbiota, reaching densities greater than 103 strains/cm2 17, enough for contamination through direct contact or flaking skin. Normal skin flora is considered one of the main origins of bacteria in indoor environments 18,19, especially in bathrooms 8 and other surfaces of the home 9, similar to the findings of the present study. CNS are normal commensal bacteria present on healthy human skin and mucous membranes; however, approximately 10% are multidrug resistant 17.
A study found that 42% of the isolates of microorganisms found on skin were resistant to erythromycin, 24.3% to tetracycline, and 13.2% to oxacillin 19, similar to the findings presented here. However, in the present study, CNS presented greater resistance, as the automated methodologies revealed the presence of the MRS phenotype in two S. hominis strains, and 17 strains resistant to beta-lactamases, macrolides, and clindamycin (S. hominis, S haemolyticos, S. cohnni, S. saprophyticus, S. epidermidis and S. warneri). he most commonly isolated phenotype profile of CNS was resistant to ampicillin, penicillin, oxacillin, and trimetroprim-sulfametoxazol, either with or without susceptibility to first-generation cephalosporins, clindamycin, erythromycin, gentamicin, and tetracycline. he present study corroborated the emergence of oxacillin-resistant and multidrug resistant CNS, which has been reported as a matter of public health concern in recent years 16.
In the present study, four strains of S. aureus were isolated from classrooms, mattresses, toys, and kitchens were sensitive to methicillin (MSSA) and the other analyzed antibiotics. Only one strain presented intermediate resistance to vancomycin. he results corroborate those of a study conducted by Scott et al. 8, which reported methicillin-sensitive S.aureus isolated from toys; however, the bacteria identified in bathrooms and kitchens were found to be resistant (MRSA). his finding is noteworthy, considering that children can acquire these bacteria through surface contact, enhancing their transmission and generating an increase in community-acquired MRSA infections.
Another relevant finding in the present study was the isolation of a multidrug-resistant strain of P. aeruginosa from bathroom surfaces. his bacterium has been reported to be a multidrug-resistant nosocomial pathogen that affects respiratory and urinary tracts, causing outbreaks in hospitals 20. However, it has also been associated with infections in people, including children, who participate in recreational activities involving exposure to sources of water in which this microorganism can survive. P. aeruginosa can be present in the water supply and sanitation systems, forming biofilm, its intrinsic resistance to various antibiotics, and high capacity to acquire genes that inhibit the action mechanisms of new antimicrobials, can incur in complications for the treatment of infections 21.
Pepperell et al., have reported that enterobacteria, especially Citrobacter and Enterobacter, presented multidrug resistance associated with the genetic recombination processes triggered by the action of an integrase that enables integrons to adhere to resistance genes. The importance of Citrobacter and similar species stems from its capacity to survive long periods because of its low level of virulence; thus, resistance genes accumulate, which can be transmitted to other more highly virulent organisms 22. Considering these studies, special mention goes to the risks found in the present investigation, such as bacteria belonging to the Enterobacteriaceae (Citrobacter, Serratia, Enterobacter, and Kluyveria) family that were 100% resistant against cephalosporins, specifically first-generation ones. This can be explained by the constant and prolonged use of antibiotics and high production of beta-lactamases.
The results of the present study did not show multidrug resistance in Enterobacteriaceae. These findings are consistent with those reported by Odhiambo et al. 23, in which most of the enteric microorganisms did not present multidrug resistance against commonly used antimicrobials, such as ampicillin, tetracycline, and cotrimoxazole. However, the authors also informed resistance against these drugs by Salmonella spp, Shigella flexineri, and Shigella spp isolates.
In the present study, bacteria such as Citrobacter, E. coli, Kuyvera, Pantoea and Enterobacter were resistant to more than three antibiotics, whereas Klebsiella and Serratia were resistant to four others. These profiles are significant, considering reports of the presence of multidrug-resistant genes in strains isolated from non-hospitalized children, indicating the transmission of these microorganisms in the community 24.
Contributions of the present research include the characterization of microorganisms and antibiotic susceptibility patterns of bacterial biota, isolated from different early childhood day care centers in Bogota. These microorganisms are communicable and generate risk of infection among children. This study points to the need to conduct interventions relative to hand hygiene and carrying out surface cleaning and disinfection protocols in child care centers. Such practices can reduce exposure of children to potentially-infectious microorganisms. Efforts must be employed to ensure healthy environments, and reduce the unnecessary prescription of antibiotics, which is not only related to the issue of multidrug resistance, but also generates burdens for health services and the resources of families.















