Introduction
On December 31, 2019, the World Health Organization issued a warning about several cases of a respiratory illness emerging from the city of Wuhan, in the Hubei province of China, with clinical manifestations similar to viral pneumonia and symptoms such as coughing, fever, and dyspnea. This newly discovered virus was named COVID-19, and it is caused by the SARS-CoV-2 virus (Chung et al., 2020).
Most people who are infected with COVID-19 experience a mild to moderate respiratory illness and may recover without requiring special treatment. Older people, and those with underlying medical problems such as cardiovascular disease, diabetes, chronic respiratory disease, and cancer are more likely to develop serious complications. An efficient way to prevent and reduce infections is by quickly detecting positive COVID-19 cases, isolating patients, and starting their treatment as soon as possible. COVID-19 spreads primarily through droplets of saliva or discharge from the nose when an infected person coughs or sneezes. There are currently no specific treatments for COVID-19 (WHO, 2020) and, as the global pandemic progresses, scientists from a wide variety of specialties play pivotal roles in developing new diagnostic, forecasting, and modeling methods. For example, Sun and Wang (2020) developed a mathematical model to characterize imported and asymptomatic patients without strict a 'quarantine', which was useful in modeling the COVID-19 epidemic. Boccaletti et al. (2020) presented an evolutionary modeling of COVID-19 using mathematical models and deep learning approaches. However, regarding diagnostic and detection methods, much of the current literature has focused on radiology techniques such as portable chest radiography (X-rays) and computed tomography (CT) scans (Chung et al., 2020; Zhou et al., 2020). For instance, Wang et al. (2021) used CT images and pre-trained deep learning models together with discriminant correlation analysis to classify COVID-19, pneumonia, tuberculosis, and healthy cases. Zhang et al. (2021) used 18-way data augmentation, a convolutional block attention module, and CT images to diagnose COVID-19 and healthy cases. However, due to infection issues related to transporting patients to CT suites, inefficiencies in CT room decontamination, and the lack of CT equipment in some parts of the world, X-rays will likely be the most common way to identify and follow up lung abnormalities including COVID-19. Additionally, depending on the air exchange rates, CT rooms may be unavailable for approximately one hour after imaging infected patients (ACR, 2020). Therefore, chest X-ray utilization for early COVID- 19 detection plays a key role in areas all over the world with limited access to reliable RT-PCR (reverse transcription polymerase chain reaction) tests for COVID-19 (Jacobi et al., 2020).
Furthermore, Artificial Intelligence (AI) methods are among the most widely used tools in the fight against the COVID-19 pandemic. Castillo and Melin (2020) proposed a model for forecasting confirmed cases and deaths in different countries based on their corresponding time series using a hybrid fuzzy-fractal approach. Particularly in the case of Mexico, Melin et al. (2020) forecasted the COVID-19 time series at the state and national levels with a multiple ensemble neural network model with fuzzy response aggregation. Castillo and Melin (2021) proposed a hybrid intelligent fuzzy fractal approach for classifying countries based on a mixture of fractal theoretical concepts and fuzzy logic to achieve an accurate classification of countries based on the complexity of their COVID-19 time series.
As of March 2021, 127 349 248 confirmed cases of COVID-19, including 2 787 593 deaths have been reported worldwide (WHO, 2021), and the numbers keep increasing. Therefore, it is critical to detect cases of COVID-19 as soon as possible in order to prevent the further spread of this pandemic, as well as to instantly treat affected patients. The need for auxiliary diagnostic tools has increased, and recent findings obtained using radiology techniques suggest that X-ray images contain significant information about COVID-19 infections. AI methods coupled with radiology techniques can be helpful for the accurate detection of this disease and can also help overcome the shortage of radiologists and experts in remote places (Ozturk et al., 2020).
AI methods, especially machine learning (Raschka and Mirjalili, 2017; SAS Institute Inc., 2018) and deep learning approaches (Beysolow II, 2017; Chen and Lin, 2014; James et al., 2013) have been successfully applied in various areas such as IoT (Yang et al., 2019), manufacturing (Lee et al., 2015), computer vision (Rosebrock, 2017), autonomous vehicles (Nguyen et al., 2018), natural language processing (Zhou et al., 2020), robotics, education (Picciano, 2012) and healthcare (Rong et al., 2020).
In healthcare, machine learning and deep learning techniques such as linear discriminant analysis, support vector machines, and feed forward neural networks have been used in several classification problems like lung disease classification (Varela-Santos and Melin, 2020), efficient segmentations of cardiac sound signals (Patidar and Pachori, 2013), classification of motor imagery electroencephalogram signals for enhancing brain-computer interfaces (Gaur et al., 2015), and the classification of normal and epileptic seizure electroencephalogram signals (Pachori et al., 2015). However, convolutional neural networks (CNNs) are the state-of-art method when it comes to image classification (Chollet, 2018) and provide better results in non-linear problems with highly dimensional spaces. CNNs have been used for signal and biomedical image processing (Wang et al., 2018), for classifying multidimensional and thermal images, and for image segmentation (Fasihi and Mikhael, 2016).
CNNs have several characteristics that justify their status as an AI and image processing tool. According to Chollet (2018), these characteristics are simplicity, scalability, and versatility. Simplicity refers to the fact that CNNs remove the need for feature engineering, replacing complex and heavy pipelines with simple and end-to-end trainable models. CNNs are scalable because they are highly suitable for parallelization on GPUs or TPUs. Additionally, CNNs can be trained by iterating over large or small batches of data, allowing them to be trained on datasets of arbitrary size. Versatility and reusability are regarded as features because, unlike many prior machine-learning approaches, CNNs can be trained on additional data without restarting from scratch, thus making them viable for continuous learning. Furthermore, trained CNNs are reusable, for example, CNNs trained for image classification can be dropped into a video processing pipeline. This allows reinvesting and reusing previous work into increasingly complex and powerful models. Additionally, new techniques and researches have recently emerged to further improve the performance of CNNs, such as the novel convolutional block attention module (Zhang et al., 2021), data augmentation techniques, new architectures and pre-trained models (He et al., 2016; Nayak et al., 2021), optimization algorithms (Kingmaand Ba, 2014), activation functions (Pedamonti, 2018), regularization techniques such as dropout (Srivastava et al., 2014) and batch normalization (Chen et al., 2019), among others.
Finally, CNNs may be deployed into devices with limited capacities when their size is appropriate, so that untrained personal use these devices to diagnose many kinds of illnesses in undeveloped areas (Rong et al., 2020).
Recently, radiology images (X-ray images) in conjunction with deep learning methods (such as CNNs) have been investigated for COVID-19 detection in order to eliminate disadvantages such as the insufficient number of RT-PCR test kits, testing costs, and the long wait for results.
Wang et al. (2020) used traditional neural networks with transfer learning from ImageNet database (Liu and Deng, 2015) to obtain a 93,3% accuracy in classifying 3 classes (COVID-19, normal, and pneumonia). Ioannis et al. (2020) developed a deep learning model using 224 confirmed COVID-19 images. Their model obtained 98,75 and 93,48% accuracy in classifying 2 and 3 classes, respectively. Sethy et al. (2020) obtained a 95,3% accuracy using 50 chest X-ray images (25 COVID-19 + 25 Non-COVID) in conjunction with a ResNet50 (He et al., 2016) pre-trained model and SVM. Ozturk et al. (2020) proposed a deep neural network using 17 convolutional layers, obtaining 98,08 and only 87,02% of accuracy in classifying 2 and 3 classes, respectively. Narin et al. (2020) obtained a 98% of accuracy using 100 chest X-ray images (50 COVID-19 + 50 Non-COVID) in conjunction with the ResNet50 (He et al., 2016) pre-trained model. Mahmud et al. (2020) used a multi-dilation convolutional neural network for automatic COVID-19 detection, achieving 97,4% and 90,3% of accuracy in classifying 2 (COVID-19 and Non-COVID-19) and 4 classes (COVID-19, normal, viral pneumonia, and bacterial pneumonia), respectively. Loey et al. (2020) used a detection model based on GAN and transfer learning to obtain 80,6, 85,2, and 99,99°% for four (COVID-19, normal, viral pneumonia, and bacterial pneumonia), three (COVID-19, normal and bacterial pneumonia) and two classes (COVID-19 and normal), respectively. Nayak et al. (2021) evaluated the effectiveness of eight pre-trained CNN models such as AlexNet, VGG-16, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50, and Inception-V3 for classification two classes (COVID-19 and normal). Varela-Santos and Melin (2021) compared traditional neural networks, feature based traditional neural networks, CNNs, support vector machines (SVM), and k-nearest neighbors to predict 2, 3, and 6 output classes, obtaining as high as 88,54% when more than 3 outputs classes were present in the data set. Chaudhary and Pachori (2020) used the Fourier-Bessel series expansion-based dyadic decomposition method to decompose an X-ray image into sub-band images that are fed into a ResNet50 pre-trained model, where ensemble CNN features are finally fed to the softmax classifier. In the study, 750 images were used (250 for pneumonia, 250 for COVID-19, and 250 for normal).
In most of these cases, the obtained results are biased and overfitted due to a huge network capacity (specially in transfer learning approaches) compared with the small number of COVID-19 images. Also, the classification scenarios are limited to only 2 or 3 classes. It should be highlighted that the larger networks will likely overfit, while simpler models are less likely to do so.
In turn, approaches based on transfer learning (Chollet 2017; Szegedy et al., 2016) used pre-trained models in datasets scarcely related to X-ray images and would have to go through a heavy fine-tuning phase to adjust the abstract representations in the models to make them relevant for the problem at hand. However, they would end up with an excess capacity compared to the dataset size. Additionally, another drawback for transfer learning approaches is that they result in very big and heavy models, thus making it difficult to deploy them on devices with limited capacities, or acquiring Internet access if these large models are deployed in the cloud, which is not always possible in remote or poor regions.
For these reasons, the contribution of this paper is:
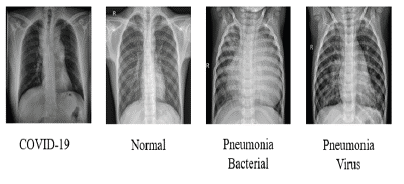
To present new models for detecting COVID-19 and other pneumonia cases using chest X-ray images and CNNs. The proposed models were developed to provide accurate diagnostics in more output classes than previous studies, covering two scenarios: binary classification (COVID-19 vs. Non-COVID) and 4-classes classification (COVID-19 vs. Normal vs. Bacterial Pneumonia vs. Viral Pneumonia). Figure 1 illustrates samples of images used for this study.
To improve accuracy in the results and prevent overfitting by following 2 actions: (1) increasing the data set size (considering more data repositories and using data augmentation techniques) while classification scenarios are balanced; and (2) adding regularization techniques such as dropout and performing automated hyperparameter optimization.
To reduce the employed network capacity and size as much as possible, thus making the final models an option to be deployed locally on devices with limited capacities and without the need for Internet access.
The impact of key hyperparameters such as batch size, learning rate, number of epochs, image resolution, activation function, number of convolutional layers, the size for each layer, and neurons in the dense layer was extensively experimented on using modern deep learning packages such as Hyperas and Hyperopt (Pumperla, 2021; Bergstra, 2021). The final proposed models achieved superior performance and required a significantly lower number of parameters compared to other studies in the literature.
Materials and Methods
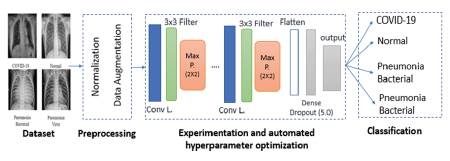
In this study, new models for detecting COVID-19 and other pneumonia cases using CNNs and X-ray images are presented. Figure 2 shows the general method used to obtain the proposed models. In general, the method consists of the following phases: consolidating the data set, preprocessing (which includes normalization and image augmentation), and experimentation and automated hyperparameter optimization.
Data set
The main challenge when applying deep learning approaches is to collect an adequate number of samples for effective training. The literature referenced in the Introduction section reveals that the earlier models are validated using a fewer number of samples, and the data in most cases is imbalanced (Nayak et al., 2021).
One of the goals of this research is to improve the accuracy and prevent overfitting by increasing the data set size, considering more data repositories and using data augmentation techniques while the classification scenarios are balanced.
For the COVID-19 class, 489 images were obtained from ISMIR (2020) and Cohen et al. (2020); 35 images were obtained from Chung (2020b), and 56 images were obtained from Chung (2020a). For the Non-COVID-19 class (Normal/Bacterial Pneumonia/Viral Pneumonia), 2 940 images were obtained from Kaggle Inc. (2020).
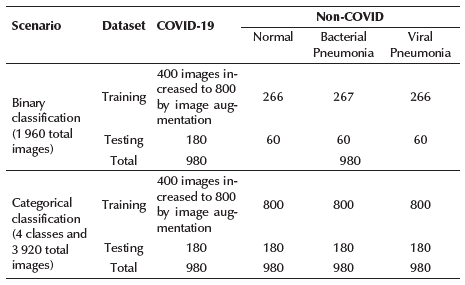
The number of images for each category is presented in Table 1. In order to avoid the issue with unbalanced data, 400 X-ray images belonging to the COVID-19 category were increased to 800 via image augmentation. This process consisted of rotating each image once in an angle between 0 and 360 degrees. Both scenarios balanced the number of images according to the available images in the COVID-19 category.
For this reason, the binary classification scenario analyzed a total of 1 960 images, 980 belonging to the COVID-19 class and 980 belonging to the Non-COVID-19 (Normal/Bacterial Pneumonia/Viral Pneumonia) class. Finally, the categorical scenario (4 classes) analyzed a total of 3 829 images, 980 for each category involved.
Experimental setup
The CNNs were developed using Python (Python Software Foundation, 2020) in conjunction with the TensorFlow(Abadi etal., 2015) and Keras (Chollet, 2015) libraries. Training and experiments were conducted on a computer equipped with an Intel Core i5-6200U processor (@ 2,40 GHz), 16 GB of RAM, 222 GB HDD, and Windows 10.
Preprocessing
Data preprocessing aims to make available raw data more suitable for neural networks (Chollet, 2018). Therefore, the collected X-ray images were preprocessed to make the training process faster and easier to implement. The images were reshaped to uniform sizes followed by value normalization (VN), which consisted of dividing the values by 255 and casting them to float type so that inputs are floating point values in the 0-1 range before processing.
Training parameters
The CNNs were trained with a backpropagation algorithm (Duchi et al., 2011) to minimize the cross-entropy loss function (binary cross-entropy for the binary scenario and categorical for the 4-classes scenario) with dropout set to 0,5 to reduce overfitting and weights updated by using the Adam optimizer (Kingma and Ba, 2014).
Each convolution layer utilized a 3 X 3 filter and an activation function after each convolution for non-linear activation, followed by a max pooling layer with 2 X 2 window and no stride (stride 1). The steps per epoch were calculated by dividing the total training sample between the corresponding batch size, and the validation steps were calculated dividing the total validation sample between the testing batch size (set to 20). Accuracy was the metric used for monitoring the training and testing phases. For the categorical scenario, softmax activation was used to produce a probability distribution over the four categories in the output layer, while sigmoid activation was used for the binary scenario to normalize the probability prediction in the output layer. A dense layer was utilized before the output layer.
Experimentation and hyperparameters optimization
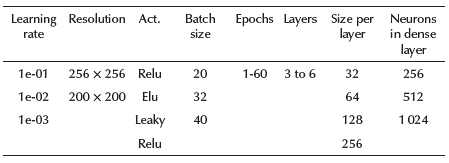
In addition to the training parameters selected in the previous section, the impact of key hyperparameters such as batch size, learning rate, number of epochs, image resolution, activation function, number of convolutional layers, the size for each layer, and neurons in the dense layer were tested.
The optimization of these hyperparameters is a difficult part that usually requires a lot of effort, time, computational resources, and extensive experimentation. But through modern deep learning packages on Python such as Hyperas (Pumperla, 2021) and Hyperopt (Bergstra, 2021), extensive experimentation and hyperparameters optimization are automated, evaluating all the options for each hyperparameter in just one execution per scenario, and automatically selecting the combination that provides the best accuracy. Just as an example, to test different values for the batch size, the code batchsize = {{choice([20,32,40])}} can be used, and these different options will be tested in the same training execution.
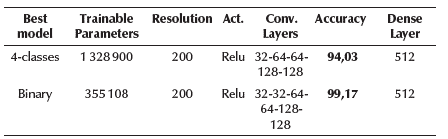
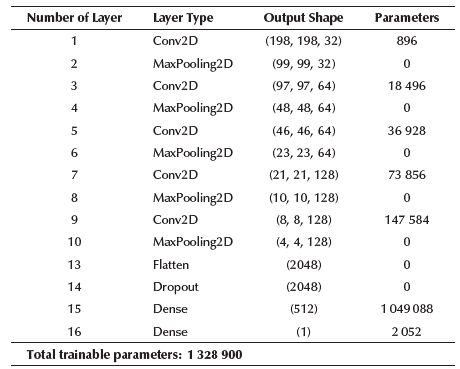
According to Mahmud et al. (2020), models that analyze COVID-19 images with a 256 X 256 resolution yield good results. Therefore, imaging was carried out in 256 X 256 and 200 X 200 resolutions (this second resolution reduces the number of trainable parameters in the models). Both of these allowed a maximum of 6 convolutional layers, so 3, 4, 5 and 6 layers were tested during 60 epochs, analyzing the accuracy, network capacity (total trainable parameters), and overfit. The tested activation functions were relu, elu, and leaky relu. The depth of the feature maps was progressively increased in the network (from 32 to 256), whereas the size of the feature maps decreased (for instance, from 198 X 198 to 1 X 1 when a 200 X 200 resolution was used). Table 2 shows a summary of the different choices tested for each hyperparameter, and Table 3 shows the best models, their accuracy, number of trainable parameters, and their selected hyperparameters after experimentation. The models were validated through cross-validation, confusion matrix and precision, recall, and F1-score values.
Results and discussion
This study presents new CNN models for detecting COVID-19 and other pneumonia cases using X-ray images in two different scenarios. These scenarios are binary classification (COVID-19 vs. Non-COVID) and 4-classes classification (COVID-19 vs. Normal vs. Bacterial Pneumonia vs. Viral Pneumonia). In both cases, a larger number of COVID-19 images was obtained to train and test the models. The classification problems were also balanced, dropout was used for regularization, and a balance was found between too much capacity and not enough network capacity through extensive experimentation. Additionally, extensive experimentation and hyperparameter optimization were automated using modern deep learning packages, and the impact of key hyperparameters was evaluated, such as batch size, learning rate, number ofepochs, image resolution, activation function, number of convolutional layers, the size for each layer, and neurons in the dense layer.
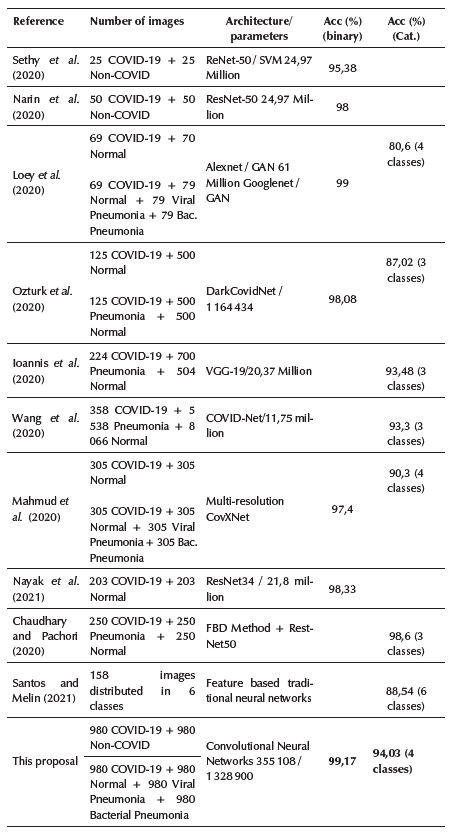
Transfer learning approaches such as the one by Wang et al. (2020) obtained a 93,3% accuracy in classifying 3 classes using a pre-trained model based on ImageNet and 11,75 million parameters. Sethy et al. (2020) obtained a 95,3% accuracy using only 50 chest X-ray images and 24,97 million parameters (ResNet50 + SVM). Narin etal. (2020) obtained a 98% accuracy using only 100 chest X-ray images and 24,97 million parameters (ResNet50). Loey et al. (2020) obtained only 80,6 and 99,99% for 4 and 2 classes using detection models based on GAN, transfer learning and 61 million parameters. Nayak et al. (2021) obtained a 98,33% accuracy in classifying 2 classes using ResNet-34 and 21,8 million of parameters. Ioannis et al. (2020) obtained 98,75 and 93,48% accuracy in classifying 2 and 3 classes, respectively, using 224 COVID-19 images, a deep learning model based on VGG-19 network, and 20,37 million parameters. Chaudhary and Pachori (2020) obtained a 98,3% accuracy classifying 3 classes using 750 total images but without specifying the network capacity, trainable parameters, or computational complexity for the final model.
In all these cases, the obtained results are biased and overfitted due to the huge network capacity (million parameters) compared with the small number of COVID-19 images. Moreover, scenarios are limited to only 2 or 3 classes. In turn, the pre-trained models are little related to X-ray images and had to go through a heavy fine-tuning phase to adjust the models to make them relevant for the problem at hand, but this resulted in models with excess capacity compared to sample size. Another drawback of these approaches is that they end up in very big and heavy models, thus making it difficult to deploy them on devices with limited capacities. People may also require Internet access to large models deployed in the cloud, which is not always possible in remote or poor regions.
Other approaches that do not use transfer learning such as Ozturk et al. (2020) obtained 98,08 and only 87,02% in classifying 2 and 3 classes using 17 convolutional layers and 1 164 million parameters. Mahmud et al. (2020) achieved 97,4 and 90,3% classifying 2 (COVID-19 and Non-COVID-19) and 4 classes (COVID-19, normal, viral pneumonia, and bacterial pneumonia), using a multi-dilation convolutional neural network. Varela-Santos and Melin (2021) compared traditional neural networks, feature based traditional neural networks, CNNs, support vector machines (SVM), and k-nearest neighbors for predicting 2, 3, and 6 output classes, obtaining at most 88,54% when more than 3 outputs classes were present in the data set. Additionally, it is not clear if some method is better than the others, and the network capacity or trainable parameters for these models are not specified.
Most of the previous studies have only used between 25 and 224 COVID-19 images, for analyzing just 2 or 3 categories. Furthermore, the hyperparameter optimization process is not performed in an exhaustive and automated way as this research does. Additionally, only Mahmud et al. (2020), Loey et al. (2020), and Santos and Melin (2021) analyzed 4 categories, obtaining an accuracy of 90,3, 80,6, and 88,54%, respectively, which can be clearly improved. In this research, 980 COVID-19 images were used (580 increased to 980 by image augmentation to avoid the unbalanced data problem) and a total of 1 960 and 3 920 images for each classification scenario, respectively, which is a significantly higher number of images. Furthermore, the network capacity for the binary scenario was adjusted to only 353 569 parameters, while the accuracy was increased to 99,17%, and, for the categorical scenario (4 classes involved), the network capacity was adjusted to only 1 328 900 parameters, while the accuracy was increased to 94,01%.
Extensive experimentation and hyperparameter optimization were automated, evaluating an appropriate set of options for each hyperparameter in just one execution per scenario and automatically selecting the combination that provided the best accuracy. The proposed models obtained superior performance, analyzed a larger number of classes, and required a significantly lower number of parameters compared to other studies in the literature (Table 4).
Table 4 Comparison of the proposed COVID-19 diagnostic models with previous works in the literature
Source: Authors
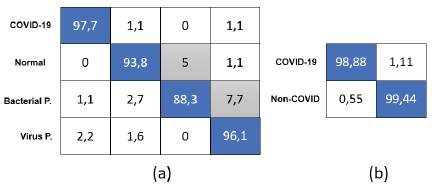
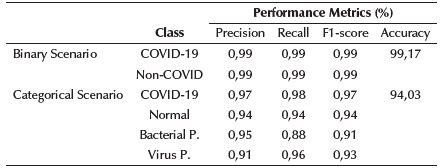
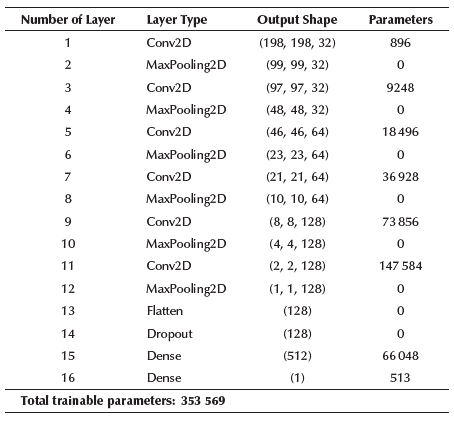
With these network sizes (number of parameters), the models are a perfect choice to be deployed locally on devices with limited capacities and without the need for Internet access. The resolution used in both cases was 200x200. For an analysis of each model, the categorical and binary confusion matrix were provided in Figures 3a and 3b, respectively, and precision, sensitivity/recall, and F1-score values are shown in Table 5. The details for the final proposed models are given in Tables 6 and 7.
The proposed models can be used for the diagnosis of COVID-19 in just a couple of seconds. These can also be widely applied because X-ray techniques are preferred to CT because they are more accessible, cheaper, and widely applied in health centers. Patients diagnosed as positive by the models can be addressed to advanced health centers for confirmation, initiating a treatment immediately. COVID-19 public images are still limited, which, in turn, is a limitation in this study; a greater number of images could help to increase the performance, especially in the categorical scenario.
Conclusions and future work
Deep learning deals with more complex and extensive data sets for which classical machine learning and statistical techniques such as Bayesian analysis and support vector machines, among others, would be impractical. Deep learning has interesting applications in healthcare areas, such as biomedicine, disease diagnostics, living assistance, biomedical information processing, among others. It has also achieved outstanding results solving problem such as image classification, speech recognition, autonomous driving, text-to-speech conversion, language translation, targeted advertising, or natural language processing. When enough labeled data is available to solve the problem, supervised learning can achieve very good performance on many tasks, for example, in image processing. For other problems, where there is insufficient labeled data, there are various deep learning techniques to enhance the model performance, such as semi-supervised and unsupervised learning with unlabeled data or transfer learning approaches using pre-trained models. A disadvantage of transfer learning approaches is that they end up in very big and heavy models, thus making it difficult to deploy them on devices with limited capacities. They may also require Internet access when they are deployed in the cloud, which is not always possible in remote or poor regions. This study presents new models for COVID-19 detection and other pneumonia cases using CNNs and X-ray images. CNNs are the state-of-art methods when it comes to image classification, and they provide better results in non-linear problems with highly dimensional spaces. The proposed CNN models were developed to provide accurate diagnostics in more output classes, while the accuracy was increased in comparison with previous studies. The proposed models prevented overfitting by following 2 actions: (1) increasing the data set size (considering more data repositories and using data augmentation techniques) while classification scenarios are balanced, (2) adding regularization techniques such as dropout and performing hyperparameter optimization.
The impact of key hyperparameters such as batch size, learning rate, number of epochs, image resolution, activation function, number of convolutional layers, the size for each layer, and neurons in the dense layer were tested. Hyperparameter optimization is a difficult part that usually requires a lot of effort, time, computational resources, and extensive experimentations. However, by using modern deep learning packages such as Hyperas and Hyperopt, extensive experimentation and hyperparameters optimization are automated, evaluating all the options for each hyperparameter in just one execution for each scenario. In previous research, the obtained results are biased and overfitted due to the huge network capacity compared with the small number of COVID-19 images, and hyperparameter optimization is done weakly and manually in just a few hyperparameters. It should be highlighted that larger networks will likely overfit, while simpler models are less likely to overfit than complex ones. The proposed models provided a satisfactory accuracy of 99,17% for binary classification (COVID-19 and Non-COVID) and 94,03% for 4-classes classification (COVID-19 vs. Normal vs. Bacterial Pneumonia vs. Viral Pneumonia), obtaining superior performance compared to other studies in the literature. Finally, the proposed models reduced the network capacity and size as much as possible, thus making the final models an option to be deployed locally on devices with limited capacities and without the need for Internet access. The developed models (available at https://github.com/carlosbelman/COVID-19-Automated-Detection-Models/) can also be deployed on a digital platform to provide immediate diagnostics.
In the future, the proposed models can be extended by incorporating more output categories and validating them by using more images. As the models were developed using Python and TensorFlow, the next step will be to deploy these models in devices with limited capacities, using Android and TensorFlow Lite, or through hybrid mobile applications using platforms such as Ionic and TensorFlow JS. In this way, the models can be deployed locally, fully automating the process of detection, and they will not require Internet access in the future, thus providing diagnosis in a few seconds, helping affected people and health centers immediately, or making them usable in remote regions affected by COVID-19 to overcome the lack of radiologists and experts.