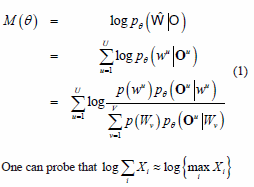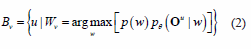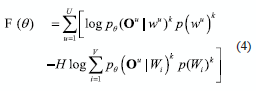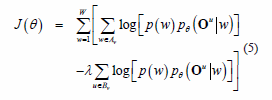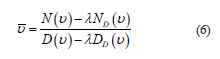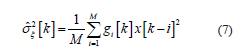Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista Facultad de Ingeniería Universidad de Antioquia
versão impressa ISSN 0120-6230versão On-line ISSN 2422-2844
Rev.fac.ing.univ. Antioquia n.55 Medellín set. 2010
On classification improvement by using an approximate discriminative hidden Markov model
Mejoramiento de la clasificación usando un modelo oculto de Markov discriminativo aproximado
Johanna Carvajal- González *, Milton Sarria-Paja, Germán Castellanos- Domínguez
Grupo de Control y Procesamiento Digital de Señales, Universidad Nacional de Colombia, Km. 7 vía al aeropuerto Campus La Nubia - Bloque W, Manizales, Colombia.
Abstract
HMMs are statistical models used in a very successful and effective form in speech recognition. However, HMM is a general model to describe the dynamic of stochastic processes; therefore it can be applied to a huge variety of biomedical signals. Usually, the HMM parameters are estimated by means of MLE (Maximum Likelihood Estimation) criterion. Nevertheless, MLE has as disadvantage that the distribution it is wanted to adjust is the distribution of each class, besides the models and/or data of other classes do not participate in the parameter re-estimation, as a result, the ML criterion is not directly related to reduce the error rate; it has led to many researchers to choice other training techniques known as discriminative training, including maximum mutual information (MMI) estimation. In this work, we carry out an EEG classification in order to compare HMM trained with both ML estimation and MMI estimation. The obtained results show a better performance in all database used.
Keywords: Hidden Markov models, discriminative training, MMI, biosignals
Resumen:
Los modelos ocultos de Markov (HMM) son modelos estadísticos usados de forma efectiva en procesamiento del habla. Aunque, siendo orientado al análisis de procesos estocásticos puede ser aplicado a una alta variedad de tareas relacionadas con el proceso e identificación con señales biomédicas. Tradicionalmente, los parámetros HMM son estimados bajo el criterio de máxima verosimilitud (entrenamiento generativo). Sin embargo, la estimación en este caso tiene como desventaja que la distribución que se quiere ajustar es la distribución de cada clase, y además los modelos y/o datos de otras clases no participan en la re-estimación de los parámetros, como consecuencia, el criterio MLE (Maximum Likelihood Estimation) no esta relacionado directamente con el objetivo de reducción de la tasa de error, lo que ha llevado a muchos investigadores a optar por técnicas de entrenamiento conocidas como entrenamiento discriminativo, en el que se encuentra la estimación de máxima información mutua. Este trabajo se realiza una comparación entre las técnicas de entrenamiento generativo y discriminativo para casos concretos de detección de patologías en señales de voz, fonocardiografía y electroencefalografía. Los resultados obtenidos muestran un mejor desempeño de la técnica discriminativa sobre la generativa en todas las bases de datos usadas.
palabras clave: Modelos ocultos de Markov, entrenamiento discriminativo, MMI, bioseñales
Introduction
Although stochastic classifiers have been employed mostly in speech recognition, their use can be extended to other biosignals tasks [1]: (i) detection of speech pathology and vocal dysfunctions [2], (ii) first and second heart sound detection and classification of different cardiac diseases by phonocardiography (PCG) [3], (iii) identification of human movements as well as pattern recognition by electroencephalography (EEG) [4]. However, the recognition performance strongly depends on the quality of the features extraction and its fit to the classifier. In conventional features extraction algorithms, discriminant analysis is one of the most promising choices for confusing classifying patterns (as it is the case of biosignal classes that manifest patterns with similar structures) where the classifier can be represented for instance by a set of discriminant functions. Nonetheless, computation of those functions requires complete knowledge of all relevant values of the probability density function (pdf) which is rarely acquired in practice, and the main goal of designing a classifier eventually is turned into by using the available training samples to estimate the class-conditional pdf P(x)|Ci) as accurately as possible. In turn, the estimation of P(x)|Ci) can be simplified by representing this density as a functional form, which consists of several adjustable parameters of a given model. Then, the estimation of the probability density becomes a problem of estimating parameters of the underlying function. One of the most common methods to overcome this issue is Maximum Likelihood Estimation (MLE) [5], that is a nondiscriminative training method, and it had become the comparison baseline in implementation of the pattern recognition systems.
Non-discriminative classifiers (referred as generative or informative classifiers [6] aim at building a model to represent the training samples for each class. Given an unknown sample, classification is carried out by choosing the model that best fit the data. Examples of non-discriminative classifiers are Hidden Markov Models (HMM) and Gaussian Mixture Models; classically, these classifiers rely on nondiscriminative training methods such as MLE, when the model of each class is trained separately by using its own samples. HMMs work well in pathology detection because the biosignal recordings are the progression of biological events that can map themselves to states. This time alignment helps in the recognition.
Most researches in HMM have focused on the estimation problem, since there is no any approach to solve analytically the model which maximizes the probability of the observation sequence [7]. Different discriminative training criteria had been proposed mainly for speech recognition. Among them the Maximum Mutual Information (MMI) and Minimum Classification Error (MCE) criteria. MMI estimation aims at finding the parameter set which maximizes the mutual information between the samples and their correct categories. MMI [8] estimation derives from the basic concept of mutual information and MCE [9], which focuses directly on minimizing the empirical classification error; both methods include the information of all classes to be recognized in the training process.
As commonly known, when samples distribution are required to be classified, these should be described by an accurate statistical model implying that the size of the training set tends to be unbounded, then the MLE training outperforms the discriminative training methods. Actually, the real data are scattered and there is a small number of records or samples [10].
This work focuses on applying a discriminative training criterion to the non-discriminative HMM classifiers with the aim of improving the recognition performance. Although HMM are used successfully and effectively in speech processing, the model can be generalized for stochastic processes and may thus be applied to a large variety of biomedical signals [11]. Since the classification of biosignals share similar characteristics with speech recognition [4], the goal of the present work is to verify whether discriminative training technique shows a better performance than generative approach (as it happens in speech pathological detection or speech recognition [12]) in training of EEG and PCG signals as well as in Voice signals. The discriminative training algorithm used in this work to estimate the HMM parameters is an approximation of the MMI objective function that is a maximization technique similar to EM algorithm, carried out by a simple modification of the standard Baum-Welch algorithm [13].
Approximated MMI algorithm
The MMI training of a model is performed over a given training set made up of the observations O = (O1,...,Ou) and their respective labels W = (w1,... wu..., wU), where each wu e (W1, Wv,...WV) and V is the classes number. Each class W1 is associated to a HMM, denoted by θ = {A,B,π}
[7].
The MMI objective function is given by [13]
One can probe that log X Xi{ « log {max Xi} [13], which is equivalent to use the MAP (Maximum a Posteriori) criterion to associate one observation with a label.
where Bv holds the indices of training set that are recognized as the class Wv, and the parameters
of each class Av = { u|wu = Wv} . Using these definitions of Av and Bv (equation 2), equation 1 is rewritten as follows:
MMI and ML can be related through of H-criterion, which is an interpolation between the MMI and ML objective functions [12]:
where k is described as a weighting exponent that usually is 1. For H = 1 this is equivalent to MMI (equation 1) and for H = 0 it is equivalent to the ML criterion [12].
Motivated by (equation 3 and 4), the following objective function is introduced, called the
approximated MMI criterion (herein, just MMI):
Note that H (equation 4) has been changed to X (equation 5). Now, it is possible describe the new re-estimation procedure for each parameter, u, in the following way:
where N(u) and D(u), referred to as accumulators, are calculated using the original training set Av. Likewise, ND(u) and Dd(u), called discriminative accumulators, are computed according to the set Bv obtained by recognition [13].
The new algorithm is developed by two following steps [13]:
Experimental setup
The experiments are performed on 3 different biosignal databases (EEG, PCG and Voice), comparing both training methods (ML and MMI). The accuracy is measured using a k-folds cross validation strategy. Namely, 10 folds have been used, splitting the 70% of the files for training classifier, and the remaining 30% for validation. The HMM topology is full connection-type, and each class is modeled by a HMM with 3 states and with diagonal covariance matrices. Besides, HMM is trained with 2, 3 y 5 Gaussian Mixtures (GM) output distributions, the number of states is fixed, it is due to the amount of degrees of freedom (Number of states, number of GM per state and the parameter λ), it makes that the number of possible combinations and the computational cost be too high, furthermore in our experiments we found if the number of states is high the algorithm does not work well, and the performance of the system is not good.
MLE is applied over the training set, and it is taken as the initial condition, the system performance is measured and it is taken as reference point, after that it is applied an iteration of Approximation and ten of maximization as suggested in [13]. Parameter λ is fixed individually according to each database. For all databases, it is found that when λ increases, values of variances and transition probabilities become negative. In this case, they are replaced by their ML values (e.g. λ = 0) [13], because the optimization method does not take into account the constrains and when they fail the model is not suitable and it has to be replaced.
EEG signals
The EEG signals are taken from Clinic of Epileptology of the University Hospital of Bonn. The database is formed by 5 sets (enumerated from A till E), each of them is composed by 100 EEG segments of a single channel that are labeled in 3 classes. The A and B sets are superficial EEG recordings (scalp) from five healthy people (normal class). The C, D and E sets refer to EEG pre-surgery diagnosis recordings as part of pathological activities (say pathological class). All EEG signal are acquired with a 128-channel system that are digitized at 173.61Hz with 12 bit resolution. We chose a single set of each class, normal and pathological, the chosen sets were A and C, respectively.
The EEG features extraction is based on a variance decimation methodology proposed in [14]. Estimation residuals of Kalman smoothing are used to compute the variance of the random process, as follow:
where x[k] is the EEG signal, g = N (2M, βσ ξ 2 [k -1]) is a Gaussian smoothing window when weight is time-variant according to the speed signal, β is an empirical constant value and M is the number samples to estimation [14].
PCG signals
The PCG database used in this work is made up of 22 de-identified adult subjects, who gave their informed consent, and underwent a medical examination. A diagnosis was carried out for each patient and the severity of the valve lesion was evaluated by cardiologists according to clinical routine. A set of 16 patients were labeled as normal, while 6 were with evidence of systolic murmur, caused by valve disorders. Besides, 8 recordings corresponding to the four traditional focuses of auscultation (mitral, tricuspid, aortic and pulmonary areas) were taken for each patient in the phase of post-expiratory and post- inspiratory apnea. Each record lasted 12 5. and was obtained from the patient standing in dorsal decubitus position. The recording time could not be extended more because patients suffering cardiac problems were unable of keeping both post-inspiratory and post-expiratory apnea for a longer period. After visual and audible inspection by cardiologists, one of the four signals was randomly picked up, taking into consideration that most of the time murmurs do not necessary show up for all focuses at once, unless they are very intense (which is an evidence of their harmfulness). An electronic stethoscope (WelchAllyn® Meditron model) is used to acquire the HS (Heart Sound) simultaneously with a standard 3-lead ECG (since the QRS complex is clearly determined, DII derivation is synchronized as a time reference). Both signals are sampled with 44.1 kHz rate. Tailored software is developed for recording, monitoring and editing the HS and ECG signals.
Application of TFR (Time Frequency Representation) to enhanced murmurs indicates that their time-frequency dynamics is far from being stationary, as it is implicitly assumed in many studies. Besides, if one demands to characterize also the dynamics of HS process, this would require a time-resolved (e.g., event- related) spectral analysis. Therefore, it is not only the spectral decomposition per se which is of interest, but rather a variety of measures derived from TFR.
Generally speaking, dynamic measures derived from TFR that have a wide acceptance for characterizing a HS [15,16] can be estimated by two methods; the ones based on computing of conditional moments of TFR, taking into account the condition of correct time and frequency marginals , and the subband methods based on filter-bank calculation.
A filter-bank applied on TFR (both Short Time Fourier Transform-STFT and Wavelet Transform- WT) and taking into account that TFR eliminates the use of smoothing window that is necessary to calculate MFCC [2], 12 MFCC are calculated with 24 filters, moreover it is applied a smoothing on the contours by using a 16-order low-pass FIR filter, with cut-off frequency of 60Hz. Choice of number of MFCC contours to be considered is made as a compromise between informativity (measured by entropy) versus consistency of estimation (measured as estimate deviation)
Voice signals
Kay-Elemetrics and UPM databases of voice disorders (described in [17]) were used to test the proposed methodology. From Kay-Elemetrics a set of 173 pathological and 53 normal speakers has been taken, the recorded material is the sustained phonation of /ah/ vowel from patients with a variety of voice pathologies: organic, neurological, and traumatic disorders [18]. UPM stores 239 pathological voices with a wide variety of organic pathologies (nodules, polyps, edemas, carcinomas, etc), and 201 normal voices. The dataset contains the sustained phonation of the /a/ Spanish vowel with a sampling rate of 50 kHz and 16-bits of resolution. Each recorded voice (observation) was uniformly windowed employing 40 ms length window with 50% of overlapping. Within each window 16 features were computed. These measures are: 12 Mel Frequency Cepstrum Coefficients (MFCC) [19], the Harmonics to Noise Ratio (HNR) [20], the Glottal to Noise Excitation Ratio (GNE) [21], the Normalized Noise Energy (NNE) [22], and the Energy of the frame, as well.
Results
EEG signals
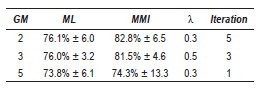
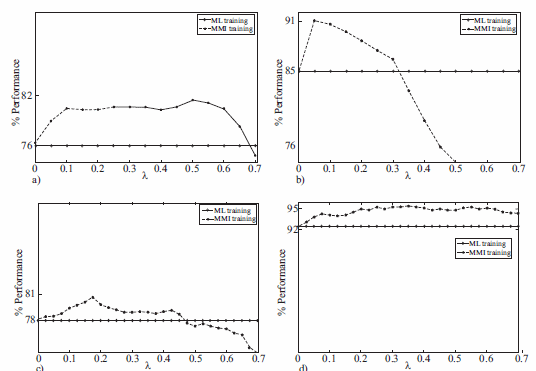
The figure 1(a) shows the recognition rate versus λ, by using 3 GM. The continuous line represents the ML baseline. In this case, best results were obtained for λ = 0.5, where the accuracy was 81.5%. In this figure, notice that when λ increases until its best performance, its behavior becomes to decrease, for this reason, λ was restricted to lower values (λ < 0.7). Similar results were found to 2 and 3 GM.
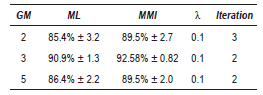
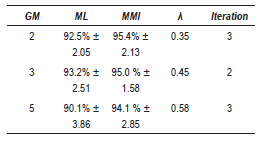
The obtained complete results with EEG signals are summarized in table 1. It is possible to see that for all GM the algorithm yielded an improvement over ML estimation. The improvement decreases while increases the number of GM, nevertheless, we can see that 3 GM performs slightly better than 2 GM. However 3 GM have less dispersion and the iteration number is lower than 2 GM.
Table 1 Best results - EEG database
The best performance for EEG signals was obtained with 2 GM, however the difference with 3 GM is approximated 1%, therefore we should taking into account the other obtained parameters as standard deviation and the iterations number and thus we can concluded that the best modeling is given for 3 GM, since both values are minor.
PCG signals
As same as in EEG signals, behavior of λ becomes to decrease, in a quicker way even to lower values than EEG case (λ < 0.35).The results on PCG database are divided in two main groups: features extraction by means of WT and STFT.
Wavelet Transform
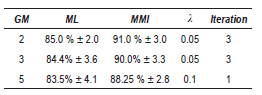
The figure 1(b) shows the recognition rate versus λ with the features obtained with WT set, by using 2 GM. Notice that when λ > 0.3 MMI performance is less than baseline ML. Similar results were found for 2 and 5 GM.
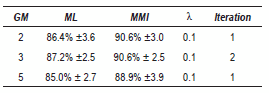
The table 2 summarized the obtained results. Better performances are always obtained to the MMI-trained model. The highest accuracy in this case was 91.0% with 2 GM.
The table 3 summarized the obtained results to the smoothed WT. In comparison with the table 2, notice that the results in MMI training are very similar, with a best performance achieve of 90.6% in the case of 2 y 3 GM, the difference between both is that the iteration number is less with the smoothed contours WT.
Table 2 Best results - PCG (contours WT)
Table 3 Best results - PCG (smoothed contours WT)
Short time frequency transform
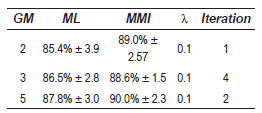
In tables 4 and 5, the results for contours STFT and smoothed contours STFT are given, respectively. The results show clearly the MMI training method always improves the recognition rate. The best performance is achieved in smoothed contours in the case of 3 GM, with the lower iteration numbers (2). All best results are obtained with λ = 0.1.
Table 4 Best results - PCG (contours STFT)
In general we can say that in PCG database the best results are obtained with λ < 0.1, and a iteration number less than or equal 4.
Table 5 Best results - PCG (smoothed contours STFT)
Voice Ssgnals
In this part we show the results to both UPM and Kay-Elemetrics databases. The λ value also was restricted and iterates by steps of 0.025.
The figure 1(c) and (d) show the recognition rate versus λ, to UPM and Kay-Elemetrics database, respectively. In this figure, we also notice that when λ increases until its best performance, its behavior becomes to decrease. Similar results were found to 2 and 3 GM. The figure for Kay- Elemetrics database is omitted because, its behavior is similar to UPM database.
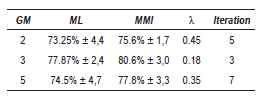
In a similar way the algorithm was tested with two voice databases (described in section III-C). The results are shown in table 6 that correspond to the evaluation of the classification system with the UPM database, it is showed that the best results are reached with 3 GM to the discriminative case when λ = 0.175 and 3 iterations are carried out, however in all cases the discriminative algorithm outperforms the ML training for λ values between 0 < λ < 0.6.
Table 6 Best results - UPM
Table 7 shows the obtained results with Kay- Elemetrics database, in this case the best results are reached when employ 2 GM were employed (λ = 0.35L=0.35) and the range for all GM of the X values is between 0 < λ < 0.7.
Table 7 Best results - Kay-Elemetrics
Despite of the accuracy achieved with the UPM database is lower than the accuracy obtained for Kay-Elemetrics database, the methodology showed to be consistent and it can be applied adequately to outperform the achieved results with a classification system based on HMM trained with ML.
The lower performance obtained with UPM database might be due to the diversity in the pathological class. This database has a large number of pathologies, hence the classes' variability is higher, and perhaps the evaluated features are not enough to model it correctly.
Figure 1 Performance vs. A. (a) EEG (b) PCG (contours WT) (c) Voice (UPM database) (d) Voice (Kay-Elemetrics database)
-------------------------------------------------------------------------------------
Conclusions
The discriminative training method for HMM based on the MMI criterion outperforms the performance of a classification system for the detection of pathologies in biosignals. This method consists on an approximation of the MMI objective function by using the similarity with the H-Criterion objective function, which is optimized by using a modified version of the BW algorithm, and it is carried out by means of an additional term that is weighted by a value λ, this term is usually referred as a discriminative accumulator. The algorithm has two major steps: Approximation, which is the derivation of the algorithm's criterion, and Maximization, which is similar to the MLE method to estimate the parameters of a HMM.
Testing of the discriminative MLE algorithm for three different types of biomedical databases (EEG, PCG and Voice) show that the operation range of λ parameter depends on the signals nature used for training. It is because the structure of randomness of the data and the source of the processes that it is wanted to model is different. Though the range turns to be different on dependence of biosignal type, suggested algorithm shows an advantage since for all considered database a better performance is achieved.
As future work, the use of other discriminative training criteria should be considered to compare between them and the training algorithm presented in this work, as well the use of contingency matrices and performance curves (ROC - curve, DET - curve) to improve the quality and clarity of the results of the validation phase.
Acknowledgments
This work was carried out under grants: 20201004208 funded by Universidad Nacional de Colombia-DIMA, "Detección de los niveles de compromiso de resonancia en niños con labio y/o paladar hendido", and "Jóvenes investigadores e Innovadores" sponsored by COLCIENCIAS and the graduate program thesis support (2009) with the project "Metodología de Entrenamiento de Modelos Ocultos de Markov Empleando Criterios Discriminativos de Gran Margen para La Detección de Patologías en Bioseñales.".
References
1. D. Novak, D. Cuesta-Frau, T. A. Ani, M. Aboy, P. Mico, L. Lhotska. "Speech Recognition Methods Applied to Biomedical Signals Processing." 26th Annual International Conference of the IEEE. San Francisco (CA). Vol. 1. 2004. pp. 118-121. [ Links ]
2. R. Solera Ureña, D. M. Iglesias, A. Gallardo, C. Peláez, A. Díaz. "Robust ASR using Support Vector Machines." Speech Communication. Vol. 49. 2007. pp. 253-267. [ Links ]
3. L. G. Gamero, R. Watrous. "Detection of the first and second heart sound using probabilistic models." Proceedings of the 25th Annual International Conference of the IEEE. Cancun (México). Vol. 3. 2003. pp. 2877-2880. [ Links ]
4. H. Lee, S. Choi. "PCA+HMM+SVM for EEG Pattern Classification." Seventh International Symposium on Signal Processing and Its Applications. Paris (France). Vol. 1. 2003. pp. 541-544. [ Links ]
5. C. M. Bishop. Neural Networks for Pattern Recognition. Ed. Oxford University Press. New York. 1995. pp. 1-508. [ Links ]
6. D. Y. Rubinstein, T. Hastie. "Discriminative vs Informative Learning". 3rd International Conference on Knowledge Discovery and Data Mining. Newport Beach (CA). 1997. pp. 49-53. [ Links ]
7. L. R. Rabiner. "A tutorial on Hidden Markov models and selected applications in speech recognition." IEEE. Vol. 77. 1989. pp. 257-285. [ Links ]
8. L. Bahl, P. Brown, P. de Souza, R. Mercer. "Maximum mutual information estimation of hidden Markov model parameters for speech recognition". Acoustics, Speech, and Signal Processing, IEEE International Conference on ICASSP. Tokyo (Japan). Vol. 11. 1986. pp. 49-52. [ Links ]
9. B. H. Juang, S. Katagiri. "Discriminative Learning for Minimum Error Classification". IEEE Transaction on Signal Processing. Vol. 40. 1992. pp. 3043-3054. [ Links ]
10. A. Nadas. "A decision theoretic formulation of a training problem in speech recognition and a comparison of training by unconditional versus conditional maximum likelihood." IEEE Trans. Acoust. Speech, Signal Processing. Vol. 31. 1983. pp. 814-817. [ Links ]
11. A. Cohen. "Hidden Markov models in biomedical signal processing". 20th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Hong Kong. Vol. 20. 1998. pp. 11451150. [ Links ]
12. Y. Normandin, S. D. Morgera. "An improved MMIE training algorithm for speaker-independent, small vocabulary, continuous speech recognition". Proceedings of the Acoustics, Speech, and Signal Processing. Vol. 1. 1991. pp. 537-540. [ Links ]
13. D. Burshtein, A. Ben-Yishai. "A Discriminative Training Algorithm for Hidden Markov Models". IEEE Transactions on Speech and Audio Processing. Vol. 12. 2004. pp. 204-217. [ Links ]
14. L. D. Avendano, J. M. Ferrero, G. Castellanos- Dominguez. "Improved Parametric Estimation of Time Frequency Representations for Cardiac Murmur Discrimination". Computers in Cardiology. Vol. 35. 2008. pp. 157-160. [ Links ]
15. H. Shino, H. Yoshida, H. Mizuta, K. Yana. "Phonocardiogram classification using time-frequency representation." 19th International Conference. IEEE/ EMBS. Chicago (IL). 1997. pp. 1636-1673. [ Links ]
16. W. Haibin, W. Jianqi, L. Guohua, Z. Guohui, N. Ansheng. "Application of adaptive time-frequency analysis in cardiac murmurs signal processing." Proceedings of the 23rd Annual International Conference of the IEEE. Istanbul (Turkey). Vol. 4. 2001. pp. 1896-1898. [ Links ]
17. G. Daza-Santacoloma, J. D. Arias-Londono, J. I. Godino, G. Castellanos-Dominguez, V. Osma, N. Saenz. "Dynamic feature extraction: an application to voice pathology detection." Intelligent Automation and Soft Computing. Vol. 15. 2009. pp. 665-680. [ Links ]
18. L. Rankinea, M. Mesbaha, B. Boashash. "IF estimation for multicomponent signals using image processing techniques in the time frequency domain." Signal Processing. Vol. 87. 2007. pp. 1234-1250. [ Links ]
19. A. Acero, X. Huang. Spoken Language Processing. Ed. Prentice Hall. New Jersey. 2001. pp. 1-1008. [ Links ]
20. G. de Krom. "A cepstrum-based technique for determining a harmonics-to-noise ratio in speech signals". Journal of Speech and Hearing Research. Vol. 36. 1993. pp. 254-266. [ Links ]
21. D. Michaelis, T. Gramms, H. W. Strube. "Glottal to Noise Excitation ratio - a new measure for describing pathological voices." Acta Acustica united with Acustica. Vol. 83. 1997. pp. 700-706. [ Links ]
22. H. Kasuya, S. Ogawa, K. Mashima, S. Ebihara. "Normalized noise energy as an acoustic measure to evaluate pathologic voice". Acoustical Society of America. Vol. 80. 1986. pp. 1329-1334. [ Links ]
(Recibido el 25 de noviembre de 2008. Aceptado el 6 de abril de 2010)
*Autor de correspondencia: teléfono: + 57 + 6 + 882 67 14, correo electrónico: johacarvajalg@gmail.com (J. Carvajal).