Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Facultad de Ingeniería Universidad de Antioquia
Print version ISSN 0120-6230On-line version ISSN 2422-2844
Rev.fac.ing.univ. Antioquia no.58 Medellín Apr./June 2011
Modeling of genetic regulatory networks in the differentiation of neural crest stem cells to sensory neurons by means of boolean networks
Modelado de redes de regulación genética en la diferenciación de células madre de la cresta neural a neuronas sensoriales a través de redes booleanas.
Jorge Marcelo Aráus Patiño1, 2, Helena Groot Restrepo2, Andrés Fernando González Barrios1*
1Grupo de Diseño de Productos y Procesos (GDPP), Department of Chemical Engineering, Universidad de los Andes. Carrera 1 E N.° 19 a 40, Edificio Mario Laserna, Bogotá, Colombia.
2Laboratorio de Genética Humana, Facultad de Ciencias, Universidad de los Andes, Carrera 1 N.° 18A -10 (M 201), Bogotá, Colombia.
Abstract
In the present study we have generated a GRN comprising the process by which neural crest stem cells develop to two types of sensory neurons (Propioceptors and Nocioceptors). We have also been able to find patterns of regulation (motifs) that act cooperatively to control such process. Surprisingly, these motifs take place in similar stages during the development of erythrocytes from hematopoietic stem cells.
Regarding the complexity of the GRN found, we then used Random Boolean Networks (RBN) for this purpose, which showed key components as well as the dynamics of the process through changes in initial conditions. Finally, the motifs were reflected in the model, suggesting insights for further studies.
Keywords: Neural crest, GRN, Boolean network, nocioceptors, propioceptors.
Resumen
En el presente estudio se ha generado una RRG que describe el proceso por el cual células madre de la cresta neural son cometidas hacia dos tipos de neuronas sensoriales (Propioceptores y Nocioceptores). Se ha encontrado también patrones de regulación (motifs) que actúan de manera cooperativa para el control de dicho proceso. Estos mismos motifs aparecen en etapas similares del desarrollo de eritrocitos a partir de células madre hematipoyéticas. Las RRG son susceptibles al modelamiento. Dada la complejidad de la RRG hallada, una red Booleana aleatoria (RBA) fue usada, la cual mostró componentes claves y la dinámica del proceso a través de cambios en las condiciones iniciales. Finalmente, los motifs fueron reflejados en el modelo, sugiriendo futuros estudios.
Palabras clave: Cresta Neural, RRG, red Booleana, nocioceptores, propioceptores.
Introduction
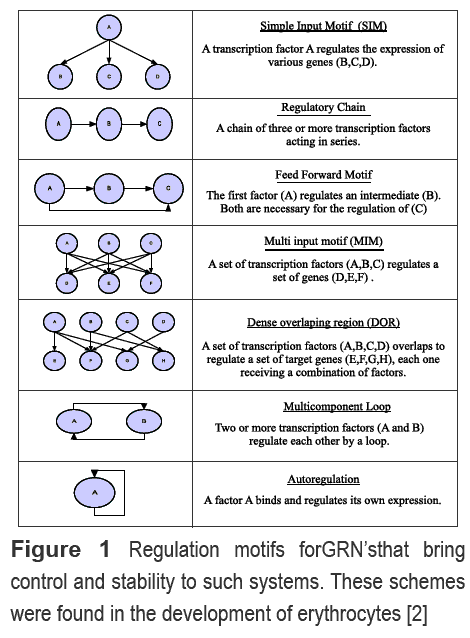
The process of development is one of the most complex phenomena found across multicellular organisms. At first glance, it appears that a coordinated combination in space and time of cisand trans elements in the DNA, transcription factors, transduction signalsand the processes that constitute the molecular biology dogmaguides undifferentiated cells to a specific fate [1], In the past decades, a great effort has been made in identifying the interaction between these molecules, generating a wealth of results. Perhaps, the best way to gather and condense these data is the generation of a genetic regulatory network (GRN) [2], which can be further characterized by patterns of transcription factor binding sites in ciselements[3], with all the molecules (intra and extracellular) involved in a spatial and temporal frame (Control logic GRN), or without taking in account these dimensions (Topological GRN) [2]. Anyway, once the network is established it is possible to find regulatory motifs en each step of the developmental process based on transcription factors. For example, in the simple input motif (SIM), a transcription activates a set of transcription factors (Figure 1A).These patterns act cooperatively and ultimately define the dynamic of the system, providing fine control and regulation [2].
Continuous and discontinuous kinetic approximations have been used to characterize GRN. To date, three types of models have been reported for this purpose, namely (i) deterministic, (ii) stochastic and (iii) Boolean models. Deterministic are based on the solution of a system of ordinary differential equations, each one expressing the kinetic interaction of one component relative to the others. Therefore, it is necessary to obtain kinetic data from molecular techniques like Real time PCR (RT-PCR)[4].Stochastic modeling is supported by the view in which genetic and biochemical networks involve the interaction of integer numbers of molecules that collide after random times, driven by Brownian motion and it requires solving the master equation based on a probabilistic function of Markovian origin [5]. Nonetheless, it is yet necessary to gather kinetic data in order to obtain the evolution of the state probability in time. On the other hand, Boolean logic based models require no kinetic parameters,hence they are suitable fordescribing large GRN. However, they are unable to provide real time dynamics as the events take place in the so called time units. These equations allow deriving gene expression time profiles based on the truth table acquired from microarrays data analysis [4].Random Boolean networks (RBN) were originally proposed for genetic modeling and they are constituted by randomly establishing the connection and the logic behind nodes. The computational resources for performing such simulations derived the attention in different alternatives such as asynchronous and temporal Boolean networks which intend to avoid the synchronicity of RBNs and probabilistic Boolean networks which incorporate the stochasticity hence avoiding the deterministic behavior once the initial conditions are established [6]
Neural crest importance could be briefly exposed as it has been recently coined "the fourth germ line"based on is novelty asits cells migrate extensively during organogenesis to generate many types of cellssuch as sensory, sympathetic and parasympathetic neurons, epidermal pigment cells and Shwann cells. In particular, the trunk neural crest migration leads to the formation of a structure named dorsal root ganglia (DRG), which is composed of two types of neurons, named propioceptorsand nocioceptors [4]. Due to its complexity, GRN comprising such complex process has not been yet reported.Here in this work we propose a Boolean based model of the differentiation of neural crest stem cells to sensory neurons that would allow to elucidate the underpinnings of its dynamics and the effect of key moleculesby in silico deletions of specific signals.
Methods
Topological GRN
Topological network was constructed based on previous reports. The underpinnings regarding precursors are yet to be uncovered due to the lack of temporal gene expression information, hence topological rather than control logic GRN was the base of our approach. Firstly, network design was grounded on Swiers et al. [2] who proposed erythrocytes development networks. Also, unique characteristics of developmental genetic networks and its hierarchical organization established by Longabaugha [3] were taken into account. Secondly, developmental process of Neural crest stem cells (NCSC) to migratory phenotypewas based on Meulemans[7], while final commitment was based on Raible[8]. Finally, specific interactions were obtained from different works [8-23].
Some types of molecules and events were obviated such as signals involved in transduction cascades and differential splicingfor the sake of simplicity. Also, in the absence of specific targets, some molecules were condensed in clusters (propioceptive battery and the combined transduction signal Wnt, Fgf and BMP). The fact that the neural crest GRN is conserved across vertebrates (Meulemans D, 2004), allows us to take results from any specie in the taxa (DanioReiro,XenopusLaevis, MusMusculus and Homo sapiens).
Boolean logic applied to a neural crest GRN
Boolean networks facilitate the analysis of GRN based on some assumptions on its structure and dynamics [4]. In the present study, Classic Boolean algorithm approach (RBN) shown by Kauffman [24] was implemented. RBN's composed of K molecules and N connections towards each component. Each molecule has an associated node, which is composed of a logic function predetermined by the behavior of the gene or protein in the network. This function is constructed by all the possible state combinations of the N connections, which in turn determine the node state. The algorithm begins with the establishment of the initial state of all the nodes (initial conditions), followed by a stage in which each molecule examines its K entry activities, consults its Boolean function and finally assumes the next state in a new discrete time. The system then traces a trajectory and it is possible that it can return to a previous state, given that the number of states is finite. In fact, the deterministic nature of the algorithm may allow the system to fall in a loop of states named attractor, which it is also a trivial property of the system [24]. Initial conditions were varied in order to determine the distribution of gene and proteins over a given discrete time, system attractors, regime, and key molecules in the network.
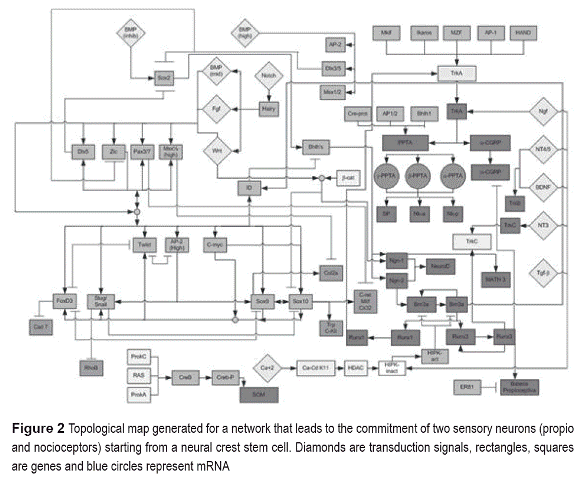
RBN was generated by analyzing interactions between genes and proteins and consequently assigning a logic function to every node (Figure 2).
Connectivity and rule matrices were established with the developed RBN and the Boolean function for each node. Due to the size of the afore mentioned network () we decided to split it in five RBN, within 8 and 9 nodes each based on criteria established elsewhere [7,8].
Results and discussion
Topological Genetic regulatory network
Figure 2 schemes the topological GRN that describes sensory neuron commitment from NCSC. These early stage cells lie at the neural plate border, captivating transduction signals from adjacent structures (Wnt, Fgf and BMP up regulated by Notch) [25]. BMP is important in the process of competence of these cells and operates in a gradient fashion over neural crest, epidermis and neural tube [8]. The low signal in the neural tube induces the transcription of Sox2, a neural expression inductor which can activate genes like N-cam and N-tubulin and is further repressed by Dlx3/5 and Dlx5.
We identified regulatory motifs at various stages of differentiation and commitment based on those reported by Swierset al. [2]. The combination of BMP (mid), Fgf and Wnt signals induce a group of transcription factors (Zic, Pax3/7, Dlx5, Msx) named neural plate border specifiers (NPBS), which mediate the influence ofthose signals and the neural crest specifiers (NCS) [7]. Two simple input motifs (SIM) (Figure 1) were identified; one conformed by BMP, AP-2, Msx1/2, Dlx3/5 and another by Dlx3/5, Sox2 and Zic. This pattern confers activation or repression of transcription programs.
With the border specified, molecular interactions permit the specification of the neural crest through its NCS's. This process is morphologically evident by an epithelium-mesenquime transition in which the NPBS induce the expression of FoxD3, Slug/ Snail, Twist, Ap-2High, Sox9, Sox10 c-myc and ID, which are known as NCS. For example, Slug/ Snail is necessary for delamination and along with FoxD3 regulates some NCS, which is known to regulate migration, delamination and induction processesby inducing genes like N-cadherin, integrins and Cadherin7. Neural crest specification is a process with high regulation and control. Two SIM (c-myc, Sox10, Twist, Slug/Snail, FoxD3 and Sox10, Twist, Slug/Snail, FoxD3), two positive multicomponent loops (Sox9 and Sox10, Sox9 and Slug/Snail), two repressive (Twist and Ap-2High, FoxD3 and Twist) and a multi input motif (MIM) between NPBS and NCS were found (Figure 1 and 2). MIM generates a great transcription program at early stages, providing spatial and temporal control via transduction signals [2]. This pattern is part of a process called antagonist cross, in which cells are competent to choose a specific fate.
Neural crest cell migration is a process in which these cells detaches from the basal lamina and are coupled to a transport system that allows the displacement to different portions of the embryo. NCS activates or repress specific neural crest factors, locomotion system and cellular support related proteins. A set of NCS (cRET, Mitf, P0, Cx32, Trp and cKit) are specific targets on genes that define the melanocyte, glia and sensory neuron genotype (Figure 2) [25]. In the same way, the coordinated action in time of signals and factors allows the progress of migration and its cease. In this regard, two SIM motifs are distinguished (Sox 10, C-ret, Mitf, Cx32, TrpcKit and Sox9, Twist, Slug/Snail, FoxD3) as well as two "feed forward" patterns (Msx1/2, Sox9, Col2a and Pax3/7, Sox 10, Cret-Mitf) that confer temporal control to GRN. Finally, a multicomponent loop between Sox9 and Sox 10 maintain the differentiated state in these cells (Figure 1 and 2).
Some of trunk neural crest cells cease its migration at a zone in which they condense to form the dorsal root ganglia (DRG). This structure receives neurotrophin transduction signals (NGF, NT4/5, BDNF and NT3), Wnt-βcatenin and TGF-β. These signals are responsible for the management of the final commitment. In such process, many regulatory motifs were found: Two autoregulation motifs (Brn3a and Runx3), one SIM (Brn3a, Runx3 and Runx1), two "feed forward" (Ngn-2, Brn3a, ID and Ngn-1,Brn3a and ID), three MIM (Ngn1/2 and NeuroD;Mkif,Ikaros, Mzf,Ap-1, Hand with TrkA; Cre-prot,Ap1/2,bhlh1 with PPTA) and four regulatory chains (Ngn- 2,Brn3a,Runx3, propioceptive battery; TrkA, PPTA and propioceptor battery; Brn3a, Runx1, TrkA; Sox2,Ngn-1 and Math3). The MIM motifs are responsible of maintain control over gene expression programs, while regulatory chain motifs provide temporal control to the system (Figure 1). In general, the validity of the GRN generated was made by comparing the presence of motifs in similar stages of differentiation on a network that depict erythrocyte commitment [2]. Surprisingly, the regulatory patterns are common for both processes, which support the view that the control of biological process is based on specific rules of regulation.
Boolean logic applied to a neural crest GRN
The topological GRN generated was divided into five Boolean networks with 8 and 9 nodes for the determination of its dynamics.Simulations were carried out at different conditions based on previously developed rule and connectivity matrices. Interestingly, each network reaches steady state during short discrete times (5 time units), presents orderly regimes and possesses one and two length attractors. In one length attractors, the steady state is unique, and the transcriptional program is activated only at the beginning. If a repressor acts when the steady state is attained, the transcriptional program is not affected. Two or more length attractors present a flexible and finer strategy for regulation, generating a feedback between transcription factors, cis and trans elements ofgenes. This generates a given pattern over time that can still be changed if a repressor is present. Hallinan [29], present a study on the constituent effect of the genes implied in the GRN, showing that of all the carried out simulations, 55% possesses a two length attractor, for networks formed with 10 nodes. The same author carries out studies on regulation motifs, in particular with those that show auto-regulative ones, showing that the best way to analyze GRN of great size is through the analysis of these control systems. With a small case of study related genes involved in cancer, the simulations showed that the analyzed motifs leads to a stationary state that behaves as an oscillator, result that was obtained here when a differentiate state was desirable.
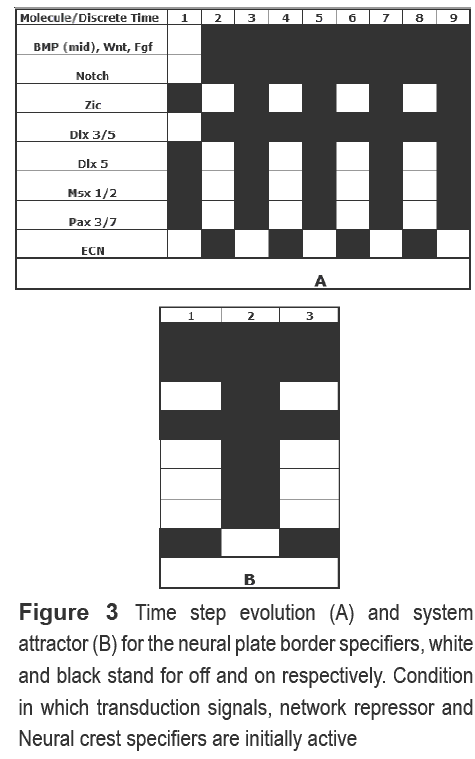
Five simulations were performed for the NPBS network. At first, we wish to determine if only the transductions signals are necessary for NPBS activation, so they were the only initial nodes active. We also wanted to know the effect of DLX3/5 and DLX5 on the system, so they were set active in two separate simulations. Two final simulations were performed in order to determine the dynamic of the differentiated state. In the first, all nodes were activated; in the second, NCS was the only node inactive. As expected, activation of the entire transduction signals lead to a specification of the border factors. Furthermore, when activating DLX5, the process is accelerated but maintaining the pattern observed when all transduction signals are activated. DLX3/5 repressor delays the specification on two discrete times. For the differentiated state simulation, we found a pattern (Figure 3) with a two length attractor, in which transduction signals are only necessary at early times for the specification of NPBS, alternating its state with NCS.
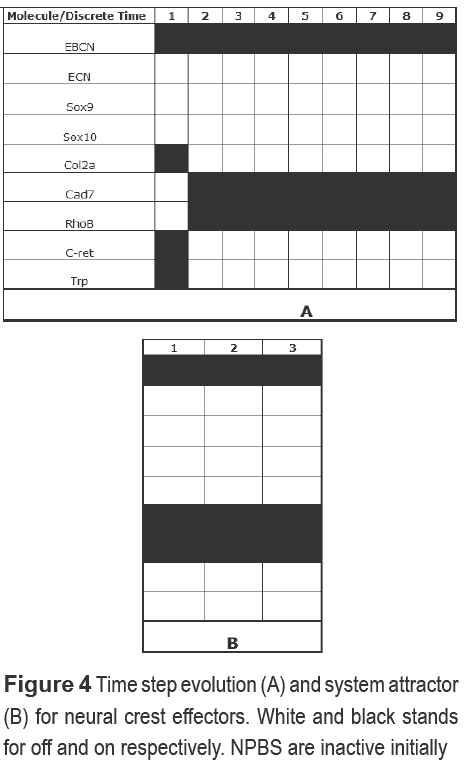
The neural crest specification was also modeled using a Boolean approximation. In this case, NPBS were grouped in a single node to determine its effect on NCS. Some motifs were not reflected (Multicomponent and SIM) partly because some molecules were excluded. We first corroborated NCS activation results obtained in the NPBS network, analyzing the system when transduction signals and NPBS are active. In order to determine if the NCS activation is caused by the activation of both we carried out a simulation for each node active finding that NCS was only activated in the presence of NPBS. NPBS and signals induced the neural crest effectors genes (NCEG) as expected, given that some NCS act repressively (Twist, FoxD3 and Slug/Snail) over NCEG (Figure 4A).
Stable activation of NCS is observed (except for Ap-2high) and a two length attractor is achieved with NCEG behaving intermittently. As in the NPBS network, a differentiated state simulation was performed, obtaining a similar pattern. Therefore, it is possible that the maintenance of early stages of differentiation occur following a global program, represented by the combination of specific states.
Cellular migrationprocess was also modeled because of its importance in the development of neural crest cells. Because of a matrix size limitation, NCS and NPBS were chosen as unique nodes. Sox9 and Sox 10 were taken as nodes, given its importance on neural crest development (Scheppers, 2002). The default state was off in every node, except for RhoB and Cad7. This state represents the initial migration process, in which the migratory phenotype is absent and RhoB and Cad7 maintain the cells attached in a epithelium fashion. NPBS alone can repress basal lamina support and maintenance related factors (Rhob and Cad7) but is unable to generate the migratory genotype. Neither can NCS, which activates Trp and the same factors that NPBS does. The migratory pattern is finally observed with a further activation of Sox9/10 and a NPBS off state. NPBS that act repressively are inactivated and those that define the neural crest are activated. Finally, all nodes excepting NPBS were activated, obtaining a similar pattern. It can be followed that temporal control is important at this stage, given that NPBS must be inactivated at a precise moment while NCS are active, allowing the migration of these cells. This fact is further represented by two "feed forward" motifs (Pax3/7, Sox 10, Cret-Mitf; Msx1/2,Sox9 and Col2a). The maintenance of the differentiated state is given by a SIM made by Sox 10 and a multicomponent loop (Sox9 and Sox10), which was also reflected on the model. In this last case, Sox 9 rescued Sox10 after two discrete times and after that, the migratory pattern was observed.
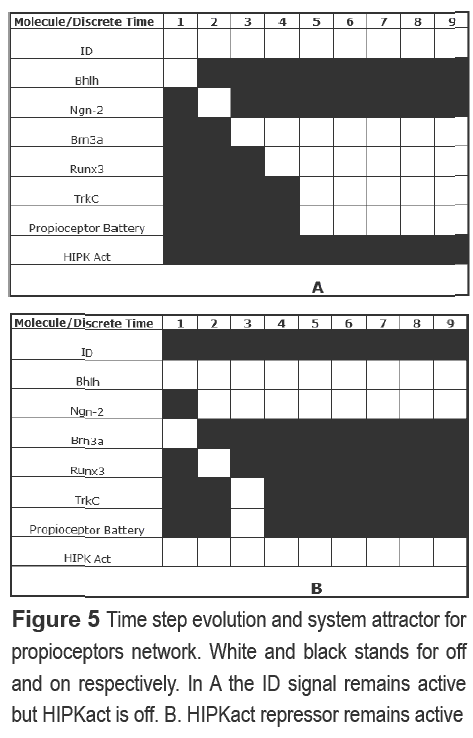
The commitment of each sensory neuron type was taken apart, generating two networks. In the propioceptor one, three general transcription factors (ID, HIPK act and Bhlh), three DRG specific factors (Ngn-2,Brn3a and Runx3), a membrane receptor (TrkC) and the propioceptive battery were included; in the nocioceptor case, we included two groups of global transcription factors ( Ikaros,Mzf,Mkif, Ap-1, Hand; Cre-prot, Bhlh-1, Ap1/2), a transduction signal (NGF), a specific transcription factor (Runx1), two related nocioceptor proteins (PPTA, α-CGRP) and the nocioceptive battery. Common points were found for both networks as for example the need of a key molecule In this regard, Brn3a is necessary for propioceptor development because (i) If its activated alone, it can promote the final genotype, (ii) its antagonist (HIPKact) repress all the network (Figure 5) and (iii) Its auto-regulative capability maintains the differentiated state
For its part, TrkA might be the molecule implicated in the definite state of nocioceptor genotype because it can activate the nocioceptive battery along with the two transcription factor nodes. However, in the absence of Runx1 and transcription factors, the molecule is unable to activate the battery. Another common point is the fact that the signals involved in the activation of the networks are needed only in a temporal manner, rather than spatially (NGF for TrkA and Bhlh for Brn3a). The dynamic in each case is reflected by a cascade fashion activation of intermediate molecules of the system (Figure 5),which also reflects the regulatory chain motif (Ngn-2, Brn3a, Runx3, propioceptive battery in propioceptors; Trka, PPTA, nocioceptive battery in nocioceptors). However, additional information about interaction between molecules is needed in the nocioceptor network.
The propioceptor specification posses nonetheless some particular characteristics that were dilucidated by the Boolean model. The Id repressive effect occurs at a temporal level over Ngn-2, given that the last is activated after some discrete times. Also, the network reflect a "feed forward" motif between Ngn-2, Brn3a, ID and two auto-regulative (Brn3a and Runx3).
Conclusions
Information gathered to date about interactions in developmental processes has made possible the construction of new GRNs. In the present study we have constructed a GRN that represents the molecular basis by which neural crest stem cells are committed to two distinct sensory neuron types. Aspects concerning the GRN presented are worth to be mentioned. The auto-renovation character of stem cells was reflected in cellular maintenance patterns in the topological map as well as in the Boolean model implemented. C-myc, implicated on cellular division process behaves in an expected way. Nonetheless, the stem cell/committed cell division was not reflected, given a lack of specific markers for each type and the fact that there is no consensus in the precursors hierarchy.
One and two length attractors were found, property related with the number of nodes and connectivity. The appearance of attractors in all cases is an indirect corroboration that the network is made in a proper way, since it is an inherent property of these systems.
References
1. S. Gilbert. Developmental Biology. 7a ed. Ed. Sinauer Associates In. Publishers. Sunderland (MA). 2006. pp. 101-137. [ Links ]
2. G. Swiers, R. Patient, M. Loose."Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification". Develop. Biol. Vol. 294. 2006. pp. 525-540. [ Links ]
3. W. J. R Longabaugha, E.H. Davidson, H. Bolouri. "Computational representation of developmental genetic regulatory networks". Develop. Biol. Vol. 283. 2005. pp. 1-16. [ Links ]
4. R. Gupta,L. E. K. Achenie."A network model for gene regulation". Comp. Chem. Eng. Vol 31. 2006. pp. 950-961. [ Links ]
5. D. Wilkinson. Stochastic modeling for Systems Biology. Ed. Chapman&Hall/CRC. Boca Raton (FL). 2006. pp. 345-370. [ Links ]
6. G. J. Hickman, P. T. Hodgman." Inference of genetic regulatory networks using boolean-network inference methods". J. Bioinform. Comput. Biol. Vol 32.2005. pp. 1-7. [ Links ]
7. D. Meulemans, M. Bronner-Fraser."Gene-Regulatory Interactions in Neural Crest Evolution and Development". Develop. Cell. Vol 7. 2004. pp. 291-299. [ Links ]
8. D. Raible."Development of the neural crest: achieving specificity in regulatory pathways". Curr. Op. Cell. Biol. Vol 18. 2006. pp. 698-703. [ Links ]
9. C. E. Krull."Segmental organization of neural crest migration". Mech. Develop. Vol. 105. 2001. pp. 37-45. [ Links ]
10. Y. Morikawa, F. D'Autreaux, M. D. Gershon,P. Cserjesi. "Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system". Develop. Biol. Vol. 307. 2007. pp. 114-126. [ Links ]
11. N. Douarin, E. Dupin. "Multipotentiality of the neural crest". Curr. Op. Gen. Develop. Vol 13. 2003. pp. 529-536. [ Links ]
12. C. Paratore, G. Brugnoli, H. Y. Lye, U. Suter, L. Sommer. "The Role of the Ets Domain Transcription Factor Erm in Modulating Differentiation of Neural Crest Cells". Develop. Biol. Vol. 250. 2002. pp. 168-180. [ Links ]
13 L. Lo, D. J.Anderson."Postmigratory Neural Crest Cells Expressing c-RET Display Restricted Developmental and proliferative capacities". Neuron. Vol. 15. 1995. pp. 527-539. [ Links ]
14. S. Eng, J. Lanier, N. Fedtsova, E. Turner. "Coordinated regulation of gene expression by Brn3a in developing sensory Ganglia". Develop. Vol. 131. 2004. pp. 3859-3870. [ Links ]
15. D. Levanon, D. Bettoun, C. Harris Cerruti, W. E.oolf, V. Negreanu,R. Eilam, Y. Bernstein, D. Goldenberg, C. Xiao, M. Fliegauf,E. Kremer, F.Otto,O. Brenner,A. Lev-Tov, Y. Groner."The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons". EMBO J. Vol. 21. 13. 2002. pp. 3454-3463. [ Links ]
16. L. Ma, J. Merenmies, L. F. Parada. "Molecular characterization of the TrkA/NGF receptor minimal enhancer reveals regulation by multiple cis elements to drive embryonic neuron expression". Develop. Vol. 127. 2000. pp. 3777-3788. [ Links ]
17. L. Ma, L. Lei, S. R. Eng, E. Turner, L. Parada. "Brn3a regulation of TrkA/NGF receptor expression in developing sensory neurons". Develop. Vol. 130. 2003. pp. 3525-3534. [ Links ]
18. L. Lei, L. Ma, S. Nef, T. Thai,L. Parada. "mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons". Develop. Vol. 128. 2001. pp. 1147-1158. [ Links ]
19. D. Levanona, O. Brenner, V. Negreaunu, D. Bettoun, R. Eilam, J. Lotem, U. Gat, F. Otto, N. Speck,Y. Grone."Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates nonredundant functions during mouse embryogenesis". Mech. Develop. Vol. 109. 2001. pp. 413-417. [ Links ]
20. A. Wiggins, G. Wei, E. Doxakis, C. Wong, A. Tang, K. Zang, E. Luo, R. L. Neve, L. F. Reichardt, E. Huang. "Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival". J. Cell Biol. Vol. 167. 2004. pp. 257-267. [ Links ]
21. H. Y. Lee, M. Kleber, L. Hari, V. Brault, U. Suter, M. M. Taketo, R. Kemler, L. Sommer."Instructive Role of Wnt/ β-Catenin in Sensory Fate Specification in Neural Crest Stem Cells". Science. Vol. 303. 2004. pp. 1020-1023. [ Links ]
22. P. White, S. J. Morrison, K. Orimito, C. Kubu, J. Verdi, D. J. Anderson. "Neural Crest Stem Cells Undergo Cell-Intrinsic Developmental Changes in Sensitivity to Instructive Differentiation Signals". Neuron. Vol. 29. 2001. pp. 57-71. [ Links ]
23. S. A. Kauffman. "Proposal for using the ensemble approach to understand genetic regulatory networks". J. Theor. Biol. Vol. 230. 2004. pp. 581-590. [ Links ]
24. S. Scott. Sensory Neurons. Ed. Oxford University. New York. 1992. pp. 45-67. [ Links ]
25. G. E. Schepers, R. D. Teasdale, P. Koopman. "Twenty Pairs of Sox: Extent, homology and nomenclature of mouse and human Sox transcription factors". Develop. Cell. Vol. 3. 2002. pp. 167-170. [ Links ]
(Recibido el 29 de septiembre de 2009. Aceptado el 1 de diciembre de 2010)
*Autor de correspondencia: teléfono: + 57 + 1 + 339 49 49 Ext. 3094, correo electrónico: andgonza@uniandes.edu.co (A. González)