Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista Facultad de Ingeniería Universidad de Antioquia
versão impressa ISSN 0120-6230
Rev.fac.ing.univ. Antioquia no.79 Medellín abr./jun. 2016
https://doi.org/10.17533/udea.redin.n79a04
ARTÍCULO ORIGINAL
DOI: 10.17533/udea.redin.n79a04
Automatic segmentation of lizard spots using an active contour model
Segmentación automática de manchas en lagartos usando un modelo de contornos activos
Jhony Heriberto Giraldo-Zuluaga1*, Augusto Enrique Salazar-Jiménez2,3
1Grupo de Electrónica de Potencia, Automatización y Robótica (GEPAR), Facultad de Ingeniería, Universidad de Antioquia. Calle 67 # 53- 108. A. A. 1226. Medellín, Colombia.
2Departamento de Ingeniería Electrónica y de Telecomunicaciones, Facultad de Ingeniería, Universidad de Antioquia. Calle 67 # 53- 108. A. A. 1226. Medellín, Colombia.
3Grupo de Automática, Electrónica y Ciencias Computacionales (AEyCC), Facultad de Ingenierías, Instituto Tecnológico Metropolitano. Calle 54A # 30-01. C. P. 050013. Medellín, Colombia.
* Corresponding author: Jhony Heriberto Giraldo Zuluaga, e-mail: heriberto.giraldo@udea.edu.co, jhonygiraldoz@gmail.com
DOI: 10.17533/udea.redin.n79a04
(Received May 05, 2015; accepted January 20, 2016)
ABSTRACT
Animal biometrics is a challenging task. In the literature, many algorithms have been used, e.g. penguin chest recognition, elephant ears recognition and leopard stripes pattern recognition, but to use technology to a large extent in this area of research, still a lot of work has to be done. One important target in animal biometrics is to automate the segmentation process, so in this paper we propose a segmentation algorithm for extracting the spots of Diploglossus millepunctatus, an endangered lizard species. The automatic segmentation is achieved with a combination of preprocessing, active contours and morphology. The parameters of each stage of the segmentation algorithm are found using an optimization procedure, which is guided by the ground truth. The results show that automatic segmentation of spots is possible. A 78.37 % of correct segmentation in average is reached.
Keywords: Diploglossus millepunctatus, active contours, gamma correction, morphological filters, spots segmentation
RESUMEN
La biometría en animales es una tarea desafiante. En la literatura muchos algoritmos se han utilizado, como, por ejemplo, el reconocimiento de los pechos en pingüinos, el reconocimiento de las orejas en elefantes y el reconocimiento de los patrones de rayas en leopardos. No cabe duda que aún hay mucho trabajo para hacer un uso masivo de la tecnología. En este artículo proponemos un algoritmo de segmentación para extraer manchas de la especie de lagartos Diploglossus millepunctatus, amenazada por la actividad humana. La segmentación automática ha sido lograda con una combinación de preprocesamiento, contornos activos y morfología. Los parámetros de cada etapa del algoritmo de segmentación han sido optimizados usando imágenes de referencia como objetivo. Los resultados muestran que la segmentación automática de manchas es posible. Un 78,37% de segmentación correcta en promedio es alcanzado.
Palabras clave: Diploglossus millepunctatus, contornos activos, corrección gamma, filtrado morfológico, segmentación de manchas
1. Introduction
Biometric identification in humans has been treated with digital signal processing aiming at recognizing hands, faces, voice, eyes, DNA, heart sound, ethnicity, scars, marks and tattoos [1]. Each topic has been extensively covered by techniques such as features extraction and machine learning. This investigation field is applied to surveillance and security systems, database extraction, tracking, and others.
The biometric methods of animal identification are application dependent, because each animal species has unique characteristics. We can search for characteristics to distinguish one individual from another one in the same species. In the literature, there are some examples, e.g. multi-curve matching of the elephants' ears is used for identifying elephants [2]. Another example is monitoring populations of penguins using their chest spots [3]. Animal biometrics is an emerging research field that combines pattern recognition, ecology and information sciences [4].
The biometric recognition of lizards is little reported in the literature, one example is the Pygmy bluetongue lizard. This lizard was identified by signature curves in [5]. The aforementioned dotted lizard (Diploglossus millepunctatus) is a lizard species native to Malpelo Island, located in the Colombian Pacific. The identification of this animal species is important, because it is in danger of extinction due to human activities [6]. To our knowledge, there is no non-invasive biometric identification method in the literature to distinguish individuals of Diploglossus millepunctatus. The method proposed here is non-invasive and based on image processing. Our algorithm includes segmentation and classification steps to identify each lizard. Various problems on spot identification can occur due to lighting variation [7] or problems due to perspective [8] that are discussed in this paper. The idea is to use segment spot patterns of Diploglossus millepunctatus to solve the named problems. In our method, a gamma correction algorithm was applied to get rid of lighting problems. Problems due to perspective were addressed by a deformable active contour model.
Some algorithms avoid the segmentation part using computer aided algorithm [7-9]; in this paper, we propose an automatic algorithm that solves the segmentation problem by dividing the segmentation into two threads. Firstly, the algorithm is applied to the image without preprocessing, with the aim to extract the darkest spots. The second thread is a gamma correction, and then the algorithm is applied to the image, with the aim to extract the lightest spots. Each thread consists of three stages. First, preprocessing that prepares the image for the active contours iteration. Second, active contours, which are not training but their parameters are tuned by an optimization algorithm; and third, deletion of atypical images via a morphological operation.
The paper is organized as follows. Section 2 shows related works. Section 3 describes the active contour method, the segmentation and the optimization algorithms. Section 4 presents the experimental framework. Section 5 exhibits the experimental results and Section 6 shows the conclusion and future works.
2. Related works
Spots segmentation has been used for medical purposes. One of the most important applications is disease diagnosis [10]. For disease diagnosis some methods are found in the literature. [11] uses Bayesian classification combined with Markov Random Field. [12] uses a watershed segmentation algorithm and [13] an active contours algorithm. The principal target of each algorithm is similar to our proposed method, but those algorithms have been used for medical purposes. The algorithm presented in this paper is used for biological purposes. Our images have illumination problems and the image is deformed by the perspective, thus the results of the segmentation of images for medical purposes and our algorithm cannot be numerically comparable. Figure 1 shows the Diploglossus millepunctatus. As can be seen, the lizard's skin is covered by a natural oily substance that gives it an unavoidable shine. The idea of this paper is to extract some lizard scales and segment the spots that are on the scales. The lizard's scales represent sets of spots that are limited by grooves.
3. Method
This section explains the methods used in this paper. First, the active contours method is explained, then the segmentation algorithm used is elucidated; finally, the optimization technique for finding the best parameters for the segmentation is explained.
3.1. Active contours
An active contours or snake is an energy-minimizing spline, guided by external constraint forces and influenced by image forces that pull it towards features such as lines and edges [14]. Active contours take an initial condition that is automatic or set up by the user.
The essential idea of active contours is to take a feature map 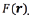 The snake is a deformable curve
The snake is a deformable curve 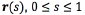 can slither on
can slither on 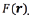 Equilibrium equations,
Equilibrium equations, 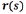 tend to cling to high responses of
tend to cling to high responses of 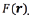 The equilibrium equation is shown in Eq. (1). In this equation, the tendency to maximize
The equilibrium equation is shown in Eq. (1). In this equation, the tendency to maximize 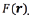 is formalized as the external energy. The counterbalance of the external energy is known as internal energy, which tends to preserve the smoothness of the curve [15]. The coefficients
is formalized as the external energy. The counterbalance of the external energy is known as internal energy, which tends to preserve the smoothness of the curve [15]. The coefficients 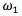 and
and 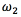 in the Eq. (1), which must be positive, are the restoring forces associated with the elasticity and stiffness of the snake, respectively.
in the Eq. (1), which must be positive, are the restoring forces associated with the elasticity and stiffness of the snake, respectively.
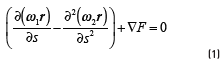
Active contours have the advantage that they do not need any previous training algorithm. Eq. (1) was solved with the Chan-Vese method [16].
3.2. Segmentation algorithm
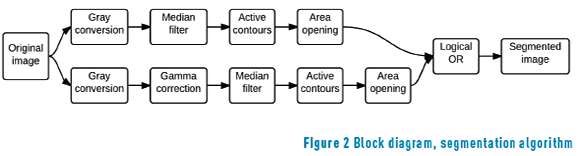
The segmentation algorithm is divided into two threads. One of the threads consists of four stages; the other thread consists of five stages. The stages are gray conversion, median filter, gamma correction, active contours and area opening. This procedure is shown in Figure 2. One thread performs segmentation without gamma correction to extract spots in the dark region; the other thread performs segmentation with gamma correction to extract spots in the bright region.
The first thread (dark region) consists of a color space transformation that is a linear combination of the original space Red Green and Blue (RGB) to result in gray space, a median filter to homogenize the region, the principal active contour iterations, and finally the area opening for deleting atypical spots. In this section, the image is processed without luminance correction, the idea is to extract spots in the darkest regions.
The gray intensity value of each pixel from RGB to gray space is defined as a weighted sum of three linear values. The luminance Y in terms of CIE 1931 is given by Eq. (2) [17], where R, G and B are the level of red, green and blue, respectively.
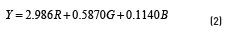
The second thread (bright region) uses the same color space transformation as the first thread, but, additionally, a decoding gamma correction. In this stage, the image is processed with a nonlinearity operation to extract the spots that are in the bright regions.
Gamma correction is a non-linear operation called power-law. Eq. (3) shows the power-law of the gamma correction [18], where 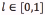 denotes the image pixel intensity, c is a constant, in common cases c = 1, and γ denotes the gamma constant. The value of γ is found by optimization techniques.
denotes the image pixel intensity, c is a constant, in common cases c = 1, and γ denotes the gamma constant. The value of γ is found by optimization techniques.

Let α be the percentage of the area opening 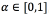 , the value of
, the value of 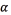 is found by optimization techniques. Let w and h be the image dimensions and let
is found by optimization techniques. Let w and h be the image dimensions and let 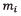 be the area of the spot i. Eq. (4) shows the area opening algorithm.
be the area of the spot i. Eq. (4) shows the area opening algorithm.
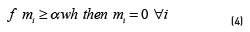
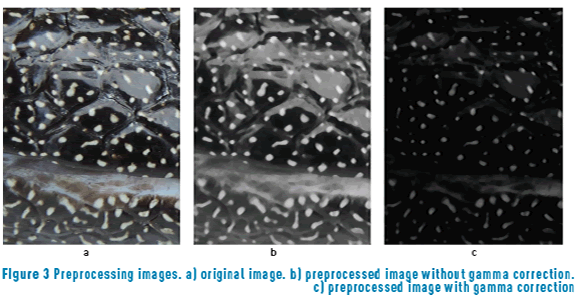
Figure 3 shows an example of the preprocessing stage. Figure 3(a) shows one original image. Figure 3(b) shows the first thread (dark region) of image preprocessing. As can be seen, the luminance problem is not resolved, because the idea is to extract the spots in the dark region. Figure 3(c) shows the second thread (bright region) of image preprocessing. This figure shows that the spots in dark regions are overshadowed. However, the spots in bright regions are not overshadowed, so it is possible to extract the spots that are in bright regions with this preprocessing procedure.
The result of active contours is binary images for each thread. Finally the two threads are merged by a logical or.
3.3. Optimization algorithm
It is necessary to choose the best parameters for the segmentation algorithm. To reach this objective, an exhaustive search was run. First we established a ground truth of some lizard scales under various conditions.
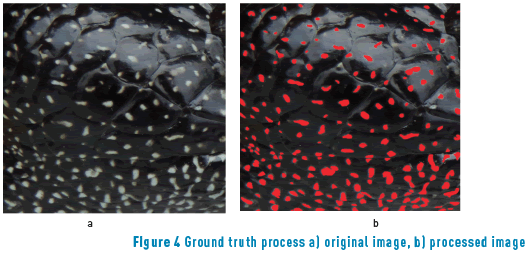
In computer vision the ground truth (GT) plays an important role in the evaluation process. The GT is important for the development of new algorithms, to compare different algorithms, and to evaluate performance, accuracy and reliability [19]. For instance, in this paper, Figure 4(a) shows one original image of the database and Figure 4(b) shows its corresponding ground truth image. The ground truth validation is used in this paper to help the optimization algorithm that is explained below.
Let X be the 2x2 confusion matrix (background and foreground) between the ground truth image and the segmented image, X11is the percentage of the background that was segmented as background, X12 is the percentage of the foreground that was segmented as background, X21 is the percentage of background that was segmented as foreground and X22is the percentage of the foreground that was segmented as foreground.
For future work, the idea is to prevent the loss of information, which means that all spots should be present in the segmented image. The objective function of the optimization algorithm is shown in Eq. (5). The aim is to maximize the percentage of correct segmentation. The confusion matrix was extracted comparing the ground truth image and the segmented image.
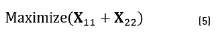
The restrictions of the algorithm are shown in Eq. (6) where γ is the gamma correction factor; ρ is the iteration number in active contours and α is the percentage in the area opening. With this algorithm, the best parameters of the segmentation were found.
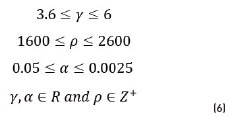
4. Experimental framework
This section explains the database conformation and describes the executed experiments. First, the optimization algorithm was executed, followed by the segmentation algorithm.
4.1. Database
The database consists of 20 samples (lizards). The images were taken under controlled conditions. 20 pieces of the original images were extracted to apply the optimization algorithm. These pieces of the original images have a resolution of 700x700 pixels, and basically are lizard scales with some spots. The database is limited, because the ground truth method is a demanding process. This is because an expert has to manually segment each image in the database.
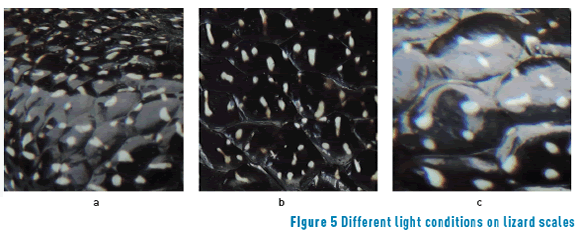
There are three kinds of light exposition lizard scales: normal (images with normal conditions of luminance), ideal (images without luminance exposure), and hard exposed images (images with high exposures to light). Figure 5 shows each kind of light condition that is present in the database.
4.2. Testing
First, the optimization algorithm was run on 20 images with combinations of all restricted parameters. For each image, the segmented algorithm was applied with 546 combinations of parameters. The 546 combinations result from 13 selected discrete values of 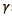 : six selected discrete values of
: six selected discrete values of 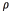 , and seven selected discrete values of
, and seven selected discrete values of 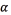 . The discrete values were selected with the experience. For each combination of parameters, the confusion matrix was extracted and the best parameter for each image was saved. The best parameters were selected with respect to the objective function of the optimization algorithm. For the selected parameters, the median was applied.
. The discrete values were selected with the experience. For each combination of parameters, the confusion matrix was extracted and the best parameter for each image was saved. The best parameters were selected with respect to the objective function of the optimization algorithm. For the selected parameters, the median was applied.
The validation testing of the segmentation was run with the segmentation algorithm on 95 images of the database. The parameters of the segmentation algorithm were the parameters found by the optimization algorithm explained above. For each segmented image, the binary confusion matrix was extracted with respect to the ground truth. The median and standard deviation were calculated for all obtained confusion matrices.
5. Experimental results
This section shows the experimental results of the segmentation algorithm, followed by a discussion about the objective function of the optimization algorithm. Finally, some visual results are exhibited.
Table 1 shows the result of the segmentation algorithm. The performance of the segmentation algorithm is 78.37% that is 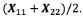
Table 2 shows the classical metric precision, recall and f-measure [20]. Each metric was extracted with the images of the validation set, and the mean and standard deviation are shown in the table.
The results of the segmentation algorithm in the validation have high standard deviation results. The classical metrics support this affirmation, where the standard deviation of each metric is close to 20%. The precision metric gives us a notion of correct spots over all supposed spots in the segmentation algorithm. The results of the classical metrics have high standard deviations, maybe due to the variability of the light exposure.
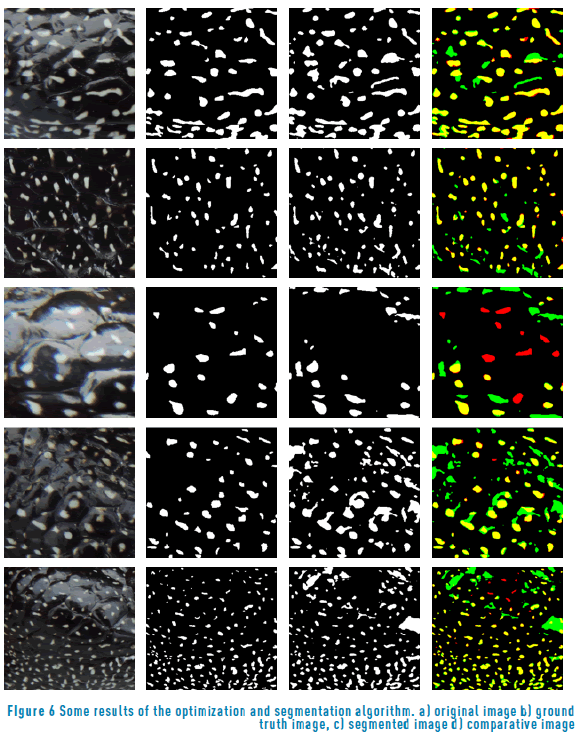
Figure 6 shows some visual results of the segmentation algorithm. Figure 6(a) shows the original image. Figure 6(b) shows the binary ground truth images. Figure 6(c) shows the segmented images; and Figure 6(d) shows the comparative image algorithm. Red-colored regions mean spots that are on the ground truth image but not on the segmented image (false negatives). Spots with yellow color mean spots that are in both, the ground truth and segmented images (true positives). Green color spots mean spots that are in the segmented image but not in the ground truth image (false positives). For a better understanding look at the digital version.
Figure 6(d) shows that the segmentation algorithm has some limitations, especially with high expose images. The limitations could be caused, because the search space shown in Eq. (6) is a reduced search space, which means that the solution given by the optimization algorithm could be a local minimum. In future work, this problem can be solved using a heuristic technique to avoid local minima.
Finally, the average running time of the segmentation algorithm was 143.9 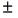 53.3 seconds per each segmented image. The experiments were carried on an Intel Core i7 3630QM with 8 gigabytes of RAM memory.
53.3 seconds per each segmented image. The experiments were carried on an Intel Core i7 3630QM with 8 gigabytes of RAM memory.
6. Conclusion and future work
We introduced an algorithm for automated segmentation of lizard spots of Diploglossus millepunctatus. The algorithm is composed of a preprocessing stage, active contours iterations and morphological filtering. The best parameters for the three algorithm stages were selected using an optimization with the objective to segment right spots and right background. The optimization algorithm needs a better design in view of practical applications, e.g. with other animals, because the processing time is already quite high. Another important aspect is that the algorithm execution, after optimization, is not as time-demanding as other segmentation algorithms in the state of the art, e.g. Markov Random Field, graph cuts and so on.
For future work, a heuristic technique will reduce the training time problem and limitations present in Eq. (6), because the optimization algorithm will be trained with more images to attain a better and more intelligent search. The next phase is the identification stage, to identify, whether a certain scale belongs to a certain lizard. In the identification stage, Procrustes Analysis, and registration algorithms will be tested.
7. References
1. J. Unar, W. Seng and A. Abbasi, ''A review of biometric technology along with trends and prospects'', Pattern Recognition, vol. 47, no. 8, pp. 2673-2688, 2014. [ Links ]
2. A. Ardovini, L. Cinque and E. Sangineto, ''Identifying elephant photos by multi-curve matching'', Pattern Recognition, vol. 41, no. 6, pp. 1867-1877, 2008. [ Links ]
3. R. Sherley, T. Burghardt, P. Barham, N. Campbell and I. Cuthill, ''Spotting the difference: towards fully-automated population monitoring of African penguins Spheniscus demersus'', Endangered Species Research, vol. 11, pp. 101-111, 2010. [ Links ]
4. H. Kühl and T. Burghardt, ''Animal biometrics: quantifying and detecting phenotypic appearance'', Trends in Ecology & Evolution, vol. 28, no. 7, pp. 432-441, 2013. [ Links ]
5. J. Li, D. Tohl, S. Randhawa, L. Shamimi and C. Bull, ''Non-invasive Lizard Identification using Signature Curves'', in TENCON, IEEE Region 10 Internationa Conference, Singapore, Singapore, 2009, pp. 1-5. [ Links ]
6. M. López, ''The lizards of Malpelo (Colombia): some topics on their ecology and threats'', Caldasia, vol. 28, no. 1, pp. 129-134, 2006. [ Links ]
7. P. Zeeuw, E. Pauwels, E. Ranguelova, D. Buonantony and S. Eckert, ''Computer assisted photo identification of Dermochelys coriacea'', in Int. Conference on Pattern Recognition (ICPR), Germany, Berlin, 2010, pp. 165-172. [ Links ]
8. M. Kelly, ''Computer-aided photograph matching in studies using individual identification: an example from Serengeti cheetahs'', Journal of Mammalogy, vol. 82, no. 2, pp. 440-449, 2001. [ Links ]
9. D. Bolger, T. Morrison, B. Vance, D. Lee and H. Farid, ''A computer‐assisted system for photographic mark–recapture analysis'', Methods in Ecology and Evolution, vol. 3, no. 5, pp. 813-822, 2012. [ Links ]
10. A. Bell, G. Herberich, D. Meyer, A. Bocking and T. Aach, ''Segmentation and detection of nuclei in silver stained cell specimens for early cancer diagnosis'', in IEEE International Conference on Image Processing (ICIP), San Antonio, USA, 2007, pp. 49-52. [ Links ]
11. J. Borovec, ''Fully automatic segmentation of stained histological cuts'', in International Student Conference on Electrical Engineering, Czech Republic, Prague, 2013, pp. 1-7. [ Links ]
12. M. Veta, A. Huisman, M. Viergever, P. Diest and J. Pluim, ''Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images'', in IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, USA, 2011, pp. 618-621. [ Links ]
13. A. Mouelhi, M. Sayadi, F. Fnaiech, K. Mrad and K. Romdhane, ''Automatic image segmentation of nuclear stained breast tissue sections using color active contour model and an improved watershed method'', Biomedical Signal Processing and Control, vol. 8, no. 5, pp. 421-436, 2013. [ Links ]
14. M. Kass, A. Witkin and D. Terzopoulos, ''Snakes: Active contour models'', International Journal of Computer Vision, vol. 1, no. 4, pp. 321-331, 1988. [ Links ]
15. A. Blake and M. Isard, Active contours, 1st ed. Cambridge, UK: Springer, 1998. [ Links ]
16. T. Chan and L. Vese, ''Active contours without edges'', IEEE Transactions on Image Processing, vol. 10, no. 2, pp. 266-277, 2001. [ Links ]
17. W. Gang et al., ''A leaf recognition algorithm for plant classification using probabilistic neural network'', in IEEE International Symposium on Signal Processing and Information Technology, Giza, Egypt, 2007, pp. 11-16. [ Links ]
18. M. Farshbaf and H. Hassanpour, ''A locally-adaptive approach for image gamma correction'', in 10th International Conference on Information Sciences Signal Processing and their Applications (ISSPA), Kuala Lumpur, Malaysia, 2010, pp. 73-76. [ Links ]
19. G. Dominguez, ''Semi-automatic generation of accurate ground truth data in video sequences'', in International Conference on Advances in Computing, Communications and Informatics (ICACCI), New Delhi, India, 2014, pp. 310-315. [ Links ]
20. D. Powers, ''Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation'', Journal of Machine Learning Technologies, vol. 2, no. 1, pp. 37-63, 2011. [ Links ]