Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Revista colombiana de Gastroenterología
versión impresa ISSN 0120-9957
Rev Col Gastroenterol vol.31 no.4 Bogotá oct./dic. 2016
A Systematic Review of Genetic Coevolution of Homo Sapiens and Helicobacter Pylori: Implications for Development of Gastric Cancer
Alix Andrea Guevara T.(1), Ángel Criollo R. (1), John Jairo Suarez O. (1), Mabel Elena Bohórquez L. MD (1),
María Magdalena Echeverry de Polanco PhD. (1)
(1) Cytogenetic Research Group, Phylogeny and Evolution of Populations in the Faculty of Sciences and Faculty of Health Sciences at the University of Tolima in Colombia
Received:Â Â Â 03-11-15Â Accepted:Â Â Â 01-11-16
Abstract
Helicobacter pylori (H. pylori) is classified as carcinogen type I for gastric cancer (GC). Although it has accompanied man for at least 116,000 years, knowledge of the evolutionary forces that modulate the role of this bacterium within the development of the spectrum of gastric diseases is still scarce. This systematic review compiles articles that report a process of coevolution process, relate host-host ancestral components, and describe H. pylori's mechanisms of adaptation to the human gastric environment in order to understand if coevolution has modulated the pathogenicity of these bacteria and the development of gastric diseases. A systematic search was carried out in MEDLINE (OvidSP), Scopus (ScienceDirect), Scielo and Tree of Science (ToS). The following search terms were used: "Stomach", "Cancer", "Neoplasms", "Ethinicity", "Evolution", "Genetics", "Ancestry" and "Helicobacter pylori", and searches were conducted in both English and Spanish. The data were filtered by one reviewer using a standard extraction form and then reviewed by another. The risk of bias and the methodological quality of the studies were evaluated using the Critical Appraisal Skills Program (CASP). Thirty-six of the total 1,584 studies found met the inclusion criteria. The most relevant factors in the development of the spectrum of diseases associated with H. pylori infection are amino acid substitutions, binding and positive selection mainly in the hypervariable regions, and disruption of the coevolution process between the bacteria and their human hosts as the result of horizontal transfer of gene segments that did not evolve with their host.
Keywords (DeCs)
Helicobacter pylori, evolution, gastric neoplasms, genetic flow.
Introduction
Gastric cancer (GC) is the fifth most common cancer in the world after lung, breast, colorectal and prostate cancer. The incidence rate among men is double that among women. In 2012, 952,000 new GC cases (6.8%)Â were estimated. Of this number, 70% come from developing countries, predominantly China (40%) GLOBOCAN (2012; http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx). In Latin America, incidence and mortality rates vary geographically and ethnically. The highest rates are found in the mountainous areas of the Pacific Coast, including Chile, Ecuador, Peru, Costa Rica and Colombia. (1, 2) Inter-population and ethnic variations in disease progression can be attributed to environmental and genetic risk factors (of either somatic or germinal origin). (3, 4)
The Helicobacter pylori bacterium coevolved with man from its origin and is very common in the intestinal microbiome. It affects more than 50% of the world's population and accounts for approximately 20% of all gastric diseases and GC. (4, 7, 12 -14) Since 1994, the International Agency for Research on Cancer (IARC) has considered it to be a Type I carcinogen and the main infectious cause of cancer in men and the second in women (after cervical cancer). (1, 8) Bacterial virulence genes (cagA, vacA and oipA) activate inflammatory pathways and produce reactive oxygen species and nitrous compounds that affect DNA stability. (8, 15, 16) For 2008, HP was responsible for approximately 89% (~ 780,000 cases) of new GC cases outside of the gastric cardia which is equivalent to 39% of the two million cases of cancer attributable to infectious agents and to 6.2% of the 12.7 million new cases reported. (10)
Since the ethnic-geographic phylogeny of this pathogen is defined with specific strains for large continental areas and geographic patterns of genetic diversity which parallel those of human diversity, (1, 3-12) some studies suggest that host-pathogen genome interactions, disruption in the coevolution process by infection with strains of ancestral origin different from the host, horizontal transfer of gene segments that have not co-evolved with hosts, and positive selection of introduced strains could generate alterations in selection for virulence and disruption of the coevolution process. This could explain the high incidence rates of GC in human populations with high genetic diversity such as Colombia which has a complex genetic mix of American, European and African origins in different proportions due to a recent process of intercontinental mixing. (17)
This systematic review aims to contribute to knowledge of the interrelationships and evolutionary forces that modulate the role of the bacterium in the development of the spectrum of gastric diseases and etiology of GC through analysis of study results describing the adaptive mechanisms of H. pylori to the human gastric environment, report host-pathogen coevolutionary processes and relate ancestral and pathogenic components of the bacterium as determinants in the development of GC.
Materials and Methods
Search Strategies
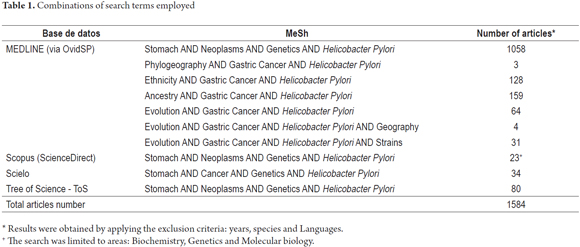
The PRISMA statement (http://www.prisma-statement.org/), MEDLINE databases (OvidSP), Scopus (ScienceDirect), Scielo and the Tree of Science - ToS (www.mytreeofscience.com) were searched using the search terms "stomach", "cancer", "neoplasms", "ethnicity", "evolution", "genetics", "ancestry" and "Helicobacter pylori" (table 1). The DOI of the articles was verified at http://www.doi.org/.
Inclusion and exclusion criteria
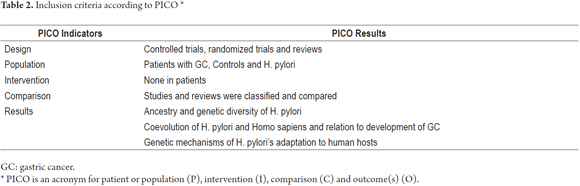
- Inclusion: studies of human populations, in English and Spanish, controlled trials, randomized trials, and reviews of: (I) genetic ancestry and diversity of H. pylori; (II) coevolution of H. pylori-Homo sapiens; (III) genetic mechanisms of adaptation of H. pylori to host as determinants in the development of GC.
- Exclusion: (I) clinical trials of drugs and vaccines; (II) comparison of treatments or diets in patients; (III) case reports; (IV) syndromes; (V) comments and editorials. Design data and study results were extracted according to the PICO acronym (Table 2).
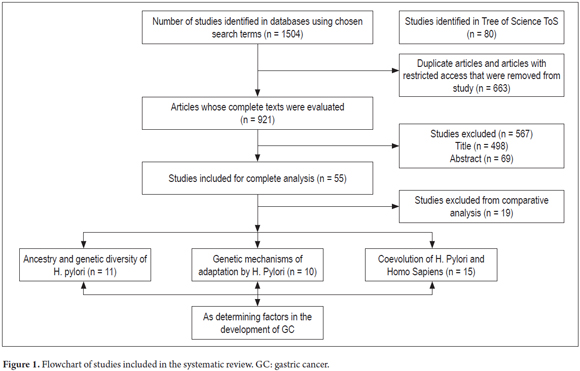
Full-text documents were independently assessed by two reviewers. Disagreements were resolved by consensus, with the participation of a third author when necessary (Figure 1).
Ancestry Distribution Map
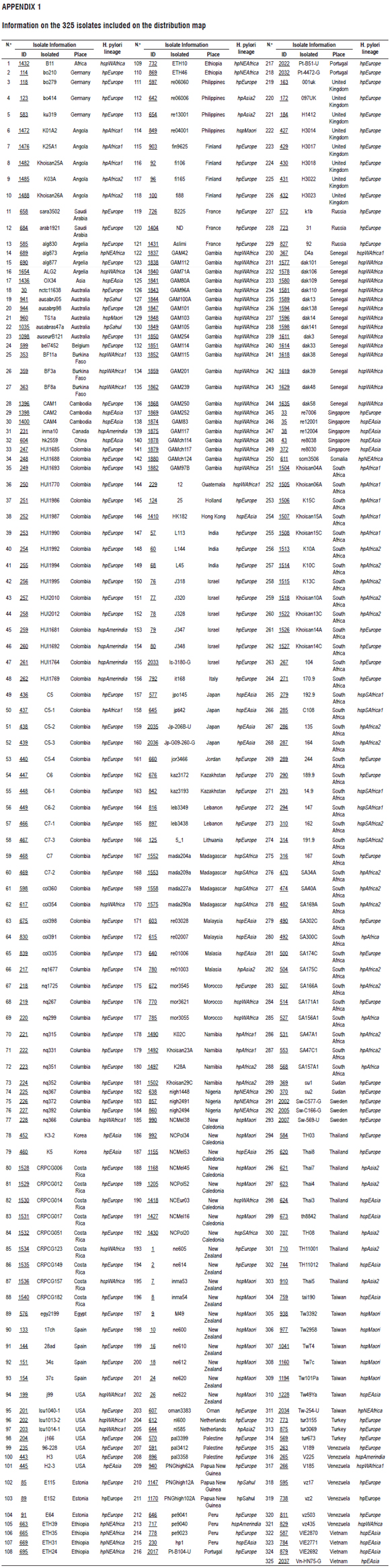
The sequences (MLST) of 7 constitutive genes (atpA, efp, mutY, PPA, trpC, ureI and yphC) of 325 isolates of H. pylori (Annex 1) that have been reported in the PubMLST database (http://pubmlst.org/) were used to map the distribution of H. pylori strains in INKSCAPE (https://inkscape.org/en/). The isolates were selected in such a way as to include all seven continents.
Quality Assessment
Quality of diagnostic studies, randomized controlled trials and reviews was assessed with the Critical Appraisal Skills Program (CASP) (http://www.casp-uk.net/casp-tools-checklists). A minimum inclusion score of 6/10 was established and then determined by two authors based on analysis of the published version.
The study was supported by publications of high methodological quality (88.9%, between 8 and 10 points), and the mean score was 8.33. No publication scored lower than 7.5.
Results
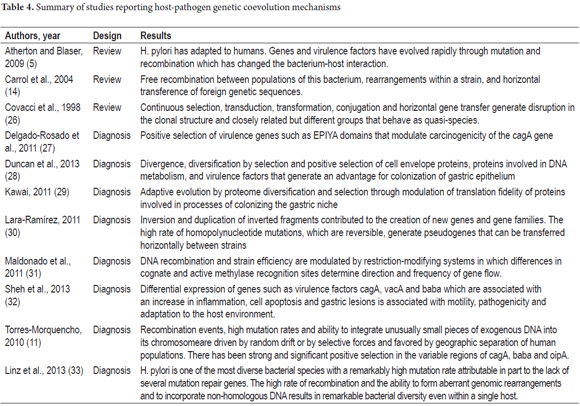
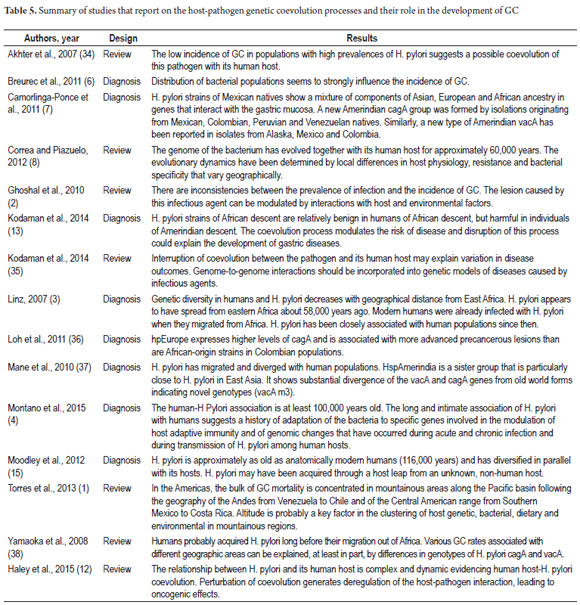
Thirty-six studies were selected for comparative analysis. Of these, 11 (30.55%) related aspects of ancestry and host-pathogen genetic diversity related to the development of gastric lesions (9, 16, 18-25) (Table 3). Ten (27.77%) reported bacterial adaptation mechanisms (5, 11, 14, 26-33) (Table 4). Finally, 15 (41.66%) related H. pylori-Homo sapiens coevolution issues as determinants in the development of GC (1-4, 6-8, 12, 13, 15, 34-38) (Table 5). Seventy-eight percent of the studies were published in journals from the USA and UK and 69.44% were published between 2010 and 2015. Diagnostic studies accounted for 69.44% and reviews accounted for 30.56%.
H. pylori infections are usually acquired orally or through fecal-oral transmission through water, food and feces during infancy. In families, transmission requires intimate contact. The presence of H. pylori varies significantly between regions. In developing countries, there are reports of higher incidence rates due to poor hygiene and water quality, contaminated food and promiscuity. (4, 6, 9, 16, 18, 39)
Epidemiological studies agree that H. pylori is a type I carcinogen and is the most important etiological agent associated with gastritis. H. pylori induces an inflammatory response which generates pre-neoplastic sequential lesions in the gastric mucosa which are associated with the development of gastroduodenal ulcers, atrophic gastritis, dysplasia, GC and MALT lymphoma. (7, 11, 12, 14, 37) H. pylori colonization may confer protection against tuberculosis through the induction of interferon antagonistic to the causative agent, mycobacterium tuberculosis. (39)
Differences among prevalences of infections and GC incidences in Africa, Malaysia, India, China, Colombia and Costa Rica could be explained by the interaction of environmental factors with host-pathogen genetic factors. In the host, these include cytokine secretion (IL-1 And IL-8) and induction of proinflammatory signals by expression of toxins, especially cagA, vacA. In the bacterium, phylogeographic origin may be important. (2, 8, 22, 36)
In Colombia, patients in the high-risk area of Nariño department who are infected with H. pylori bacteria, and who are positive for cagA and vacA s1/m1, have more severe histopathological alterations than do patients in the low risk areas on the Colombian Pacific Coast. This is due to increases in the expression of the cagA protein, increases in the expression of the enzyme spermine oxidase (SMOX), and lower rates of apoptosis in strains of European phylogeographic origin than in strains of African origin. ( 22, 36, 40)
Origin and age of association
According to 46.1% of the studies, H. pylori is one of the oldest bacteria in the intestinal microbiome. It has co-evolved with Homo sapiens from its origin and during migrations out of Africa and thus presents a geographically and ethnically defined organism with specific strains for large continental areas and geographical patterns of genetic diversity parallel to human diversity. (3, 4, 8, 12, 13, 15, 16, 35, 39) Genetic variations in H. pylori have more discriminatory power in the determination of ancient migrations in the Ladakh region of northern India and in the Pacific (Austronesian expansion) than traditional human genetic markers such as the hypervariable region (HSV1) Of mitochondrial DNA. (6) The time of association between H. pylori and its human host reported in coevolution studies ranges from approximately 60,000 years ago to approximately 116,000 years ago. (3, 4, 6-9, 13, 15, 29, 39) The oldest date for this association is 116,000 years ago. (15)
Genetic Diversity, Geographic Phylogeny and Host Adaptation Mechanisms
Eight of the studies (30.7%) agree that the diversity of the bacterium tends to decrease with increasing distance from Africa which is in line with what is observed in human populations. They also concur that the unusual intra-population genetic flexibility is due to recombination events, a high rate of mutation and the insertion of exogenous DNA fragments into the bacterial chromosome. Together they generate genomic changes that allow the evolution of areas of plasticity and regulatory mechanisms that modulate gene expression. (4, 6, 7, 11, 16, 33)
Selective pressures from the host immune response during acute and chronic infection and during the transmission of the pathogen between and among human hosts combined with polymorphisms within H. pylori strains have generated evolutionary changes in bacterial phylogeny. These occur primarily in genes involved in electron transfer, redox metabolism and DNA repair. These include the nusA transcription elongation factor; HU binding proteins which protect DNA integrity; proteins activated proteins during starvation which are necessary for survival during acid stress; genes involved in methylation patterns, an epigenetic mechanism that regulates gene expression and phenotypic plasticity; genes involved in the flagellar cascade which allow cellular motility and cell adhesion to the mucosa of the stomach which is a key factor for successful colonization of the human stomach; genes involved in metabolism of copper, cadmium, zinc, cobalt and nickel; and those involved in virulence such as cagA, vacA and oipA which can be positively selected within populations of H. pylori from different geographical origins. (2,4,6,7,11,23,29,33)
Ethnographic Evolution and Microevolution
Amino acid substitutions, binding and selective pressures primarily in the cagA, Baba, hspA, and oipA regions vary among populations of H. pylori from different geographical origins and show ethnic associations. For example, the vacA s1c region is associated with strains from East Asia whereas vacA s1b is found in strains from Spain, Portugal and Latin America. The highly polymorphic 3 âcagA region translates into different patterns of the terminal region of the protein that are differentially distributed geographically. ABD amino acid sequences flanking EPIYA (tyrosine phosphorylation) are associated with East Asian strains while the "ABC" pattern is typical of H. pylori strains in the west. ( 7, 11)
Positive selection of these genomic regions and the rate of recombination among strains should have allowed evolution of new lineages such as that of Southeast Asia (hspEAsia), consisting of Japanese and Korean genomes, and which is distinct from Amerindian, African and European lineages. (29) The vacA m3 locus, present in Amerindian populations, diverges from Old World forms which indicates that it is a recent genotype. (21, 37)
Ancestral Gene Populations
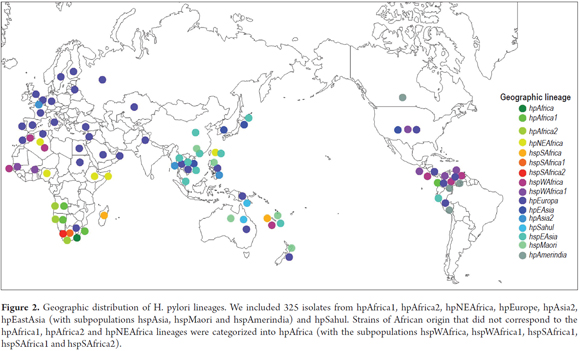
Multilocus sequence typing (MLST) was used by 61.5% of the studies to analyze genetic diversity in the sequences of 7 conserved genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC) and genes associated with virulence (vacA, cagA, hspA and oipA) in different ethnic groups. Analysis by MLST in programs such as STRUCTURE subdivided H. pylori into 7 specific populations for large geographic areas: hpEurope, hpNEAfrica, hpAfrica1, hpAfrica2, hpAsia2, hpSahul and hpEastAsia with the hspAsia, hspMaori and HspAmerindia. (3, 4, 6-9, 11-13, 16, 18, 33, 35, 37) These are derived from 6 ancestral populations: Ancestral Europe1 (AE1), Ancestral Europe2 (AE2), Ancestral East Asia, Ancestral Africa1, Ancestral Africa2 and Ancestral Sahul. (6)
According to the analyses of 325 isolates reported in the PubMLST database, there is an expansion of hpEurope strains into North Africa, Asia and the Americas and strains of hpEurope are more prevalent than are strains from other geographical origins (Figure 2 ). This could be the result of recent processes of human migration. In the Indian population, hpEurope hs an adaptive advantage in the colonization of the gastric niche and displaces strains such as hpAfrica and hpEastAsia. (25) Strains of several geographical origins (hpEurope, hspAmerindia, hpAfrica1, hspWAfrica) were reported in the departments of Bogotá and Nariño in Colombia. This is probably due to horizontal gene transfer and infection with strains from geographical origins other than those of the from native populations that were introduced by slaves and colonizers during colonization processes (Figure 2). (9, 13, 17, 18, 36)
Coevolution and GC
According to 46.1% of the articles, during acute and chronic host infection there have been adaptive events in bacterial-specific genes involved in the modulation of host adaptive immunity, reduction in the number of open reading frames, and reduction in the size of the bacterial genome. This is supported by findings from regions of Africa, Malaysia, India and Colombia where the prevalence of H. pylori infection is almost 100%, although GC incidence rates are low. (2, 4, 8, 11-13, 18, 23, 27, 29, 33)
Interactions between the host-pathogen genome and disruption in the coevolution process by infection with strains of ancestral origin other than that of the host are important in the development of GC. (8, 35). For example, in Colombia, the incidence of GC on the Pacific Coast is 6 cases/100,000 inhabitants/year while in Nariño it is 150 cases/100,000 inhabitants/year. However, the prevalence of H. pylori in these regions is similar (90%). It is interesting to note that on the Pacific Coast, the ancestry of human populations is mainly African (58%), while in Nariño it is Amerindian (67%). (13) This coincides with the fact that African strains have been shown to be benign in humans of African ancestry but harmful in individuals of Amerindian descent and indicates that coevolutionary relationships are determinant for the risk of developing GC and that colonization continues to influence the health of modern American populations.
Discussion
Humans have coevolved with the viruses and bacteria of their microbiome including human papillomavirus (HPV), hepatitis G virus, retrovirus HTLV-1 RNA and H. pylori bacteria (12, 13, 20, 26, 35). H. pylori is one of the best examples because of its adaptation to the gastric environment through modification of genes involved in the modulation of host adaptive immunity and through the evolution of host adaptation mechanisms in various human ethnic groups. (35) This has allowed for the development of a largely innocuous and potentially symbiotic infection. (4, 6, 8, 11-13, 27-29, 32, 33, 36, 37, 39)
The genetic diversity of H. pylori tends to decrease a distance from Africa increases. This is congruent with the tendency observed for genetic diversity in human populations. (3, 4, 8, 12, 13, 15, 16, 35, 39) There are approximately 1,560 genes in the constitutive genome of the bacterium while approximately 400 to 500 are specific and vary in each strain. High rates of mutation, transduction, transformation and conjugation, horizontal gene transfer in recombination events, genomic rearrangements, insertion of non-homologous DNA fragments, loss of genes during infection with multiple strains, positive selection of cell envelope proteins involved in DNA metabolism and virulence factors explain genetic diversity in the bacterial genome. Diversity can exist within the same host where the bacteria are capable of adapting to a specific gastric niche. (4, 6, 7, 11 , 12, 16, 21, 33)
Despite high variability, H. pylori shows structured ethnic and phylogeographic patterns that correlate with those of its human hosts and which result from intrafamily transmission of infections, local dispersion of single nucleotide polymorphisms due to homologous recombination and isolation resulting from distance between human populations which promotes divergence due to genetic drift and adaptation to local conditions. (4, 6, 15, 16, 21)
Coevolution of H. Pylori, Homo Sapiens and GC
Although 80% of infected individuals are asymptomatic, H. pylori is the most important etiological agent associated with gastritis and induces an active chronic inflammatory response that can affect the entire gastric mucosa. The outcome of infection is determined by the interaction of pathogen characteristics in combination with host genetic factors and environmental factors. (12, 18)
For 2008, approximately 780,000 GC cases were caused by H. pylori infection (6.2% of the 12.7 million new cases reported that year). This confirms that H. pylori is a type I carcinogen. (10)
Estudios recientes demuestran que el proceso de coevolución de H. pylori-Homo sapiens es un factor determinante que modula el desarrollo de las lesiones gástricas (3, 6, 8, 13, 15, 35, 37). La disrupción del proceso de coevolución, por la transferencia horizontal de cepas y genes que no han coevolucionado con su huésped, podría explicar en parte las tasa de incidencia de GC en poblaciones con una genética compleja, como la colombiana, que ha experimentado un proceso reciente de mezcla intercontinental entre amerindios, europeos y africanos en diferentes proporciones (13, 17). Por ende, los estudios evolutivos en esta bacteria son importantes para comprender la dinámica huésped-patógeno e identificar los procesos adaptativos y coevolutivos y las interacciones que promueven el desarrollo del espectro de enfermedades asociadas con la infección (7, 8, 13, 17, 35).
Recent studies demonstrate that the H. pylori-Homo sapiens coevolution process is a determining factor that modulates development of gastric lesions. (3, 6, 8, 13, 15, 35, 37). Disruption of the coevolution process by horizontal transfer of strains and genes that have not coevolved with their host could partly explain incidence rates of GC in populations with complex genetics. One such case is the population of Colombia which has undergone a recent process of intercontinental mixture among Amerindians, Europeans and Africans in varying. (13, 17) Evolutionary studies of this bacterium are important for understanding host-pathogen dynamics and for identifying adaptive and coevolutionary processes and interactions that promote the development of the spectrum of diseases associated with infection. (7, 8, 13, 17, 35)
Acknowledgements
The authors wish to acknowledge the work of the Research Group on Cytogenetics, Phylogeny and Evolution of Populations and the faculties of Sciences and Health Sciences of the University of Tolima for their work in the integral academic formation of students.
Authors' contributions
Alix Andrea Guevara Tique and Mabel Elena Bohórquez L. carried out the search for studies. Alix Andrea Guevara Tique was responsible for the first draft manuscript. Ángel Criollo R., John Jairo Suarez O., Mabel Elena Bohórquez L. and María Magdalena Echeverry de Polanco contributed significantly to the final version of the manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Appendix 1
Information on the 325 isolates included on the distribution map
REFERENCES
1. Torres J, Correa P, Ferreccio C, et al. Gastric cancer incidence and mortality is associated with altitude in the mountainous regions of Pacific Latin America. Cancer Causes Control. 2013;24(2):249-56. [ Links ]
2. Ghoshal UC, Chaturvedi R, Correa P. The enigma of Helicobacter pylori infection and gastric cancer. Indian J Gastroenterology. 2010;29(3):95-100. [ Links ]
3. Linz B. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915-8. [ Links ]
4. Montano V, Didelot X, Foll M, et al. Worldwide population structure, long-term demography, and local adaptation of Helicobacter pylori. Genetics. 2015;200(3):947-63. [ Links ]
5. Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119(9):2475-87. [ Links ]
6. Breurec S, Guillard B, Hem S, et al. Evolutionary history of Helicobacter pylori sequences reflect past human migrations in Southeast Asia. PloS one. 2011;6(7):e22058. [ Links ]
7. Camorlinga-Ponce M, Perez-Perez G, Gonzalez-Valencia G, et al. Helicobacter pylori genotyping from American indigenous groups shows novel Amerindian vacA and cagA alleles and Asian, African and European admixture. PloS one. 2011;6(11):e27212. [ Links ]
8. Correa P, Piazuelo MB. Evolutionary history of the Helicobacter pylori genome: implications for gastric carcinogenesis. Gut Liver. 2012;6(1):21-8. [ Links ]
9. Shiota S, Suzuki R, Matsuo Y, et al. Helicobacter pylori from gastric cancer and duodenal ulcer show same phylogeographic origin in the andean region in colombia. PloS one. 2014;9(8):e105392. [ Links ]
10. Plummer M, Franceschi S, Vignat J, et al. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136(2):487-90. [ Links ]
11. Torres-Morquecho A, Giono-Cerezo S, Camorlinga-Ponce M, etal. Evolution of bacterial genes: Evidences of positive Darwinian selection and fixation of base substitutions in virulence genes of Helicobacter pylori. Infect Genet Evol. 2010;10(6):764-76. [ Links ]
12. Haley KP, Gaddy JA. Helicobacter pylori: genomic insight into the host-pathogen interaction. Int J Genomics. 2015;2015:386905. [ Links ]
13. Kodaman N, Pazos A, Schneider BG, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci. 2014;111(4):1455-60. [ Links ]
14. Carroll IM, Khan AA, Ahmed N. Revisiting the pestilence of Helicobacter pylori: insights into geographical genomics and pathogen evolution. Infect Genet Evol. 2004;4(2):81-90. [ Links ]
15. Moodley Y, Linz B, Bond RP, et al. Age of the association between Helicobacter pylori and man. PLoS pathogens. 2012;8(5):e1002693. [ Links ]
16. Latifi-Navid S, Ghorashi SA, Siavoshi F, et al. Ethnic and geographic differentiation of Helicobacter pylori within Iran. PloS one. 2010;5(3):e9645. [ Links ]
17. Criollo Rayo AA. Caracterización molecular de la variación genética en cuatro etnias indígenas (Pijao, Paez, Embera y Zenu) y dos poblaciones mestizas de Colombia (Tolima y Córdoba) mediante marcadores del mDNA, NRY y AIMs. [Tesis de maestría]. Tolima, Colombia: Universidad del Tolima; 2013. [ Links ]
18. de Sablet T, Piazuelo MB, Shaffer CL, et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60(9):1189-95. [ Links ]
19. Devi SM, Ahmed I, Khan AA, et al. Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island. BMC Genomics. 2006;7:191. [ Links ]
20. Dominguez-Bello MG, Perez ME, Bortolini MC, et al. Amerindian Helicobacter pylori strains go extinct, as European strains expand their host range. PloS one. 2008;3(10):e3307. [ Links ]
21. Kersulyte D, Kalia A, Gilman RH, et al. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PloS one. 2010;5(11):e15076. [ Links ]
22. Martínez T, Pérez-García J, Hernández GA, et al. Características histológicas de la gastritis asociada a los genotipos cagA y vacA de Helicobacter pylori difieren en 2 zonas de riesgo opuesto para cáncer gástrico en Colombia. Rev Esp Patol. 2013;46(3):139-52. [ Links ]
23. Miftahussurur M, Sharma RP, Shrestha PK, et al. Molecular epidemiology of Helicobacter pylori infection in Nepal: specific ancestor root. PloS one. 2015;10(7):e0134216. [ Links ]
24. Yamaoka Y, Orito E, Mizokami M, et al. Helicobacter pylori in North and South America before Columbus. FEBS Letters. 2002;517(1-3):180-4. [ Links ]
25. Devi SM, Ahmed I, Francalacci P, et al. Ancestral European roots of Helicobacter pylori in India. BMC Genomics. 2007;8:184. [ Links ]
26. Covacci A, Telford JL, Del Giudice G, etal. Helicobacter pylori virulence and genetic geography. Science. 1999;284(5418):1328-33. [ Links ]
27. Delgado-Rosado G, Dominguez-Bello MG, Massey SE. Positive selection on a bacterial oncoprotein associated with gastric cancer. Gut Pathogens. 2011;3(1):1-10. [ Links ]
28. Duncan SS, Valk PL, McClain MS, et al. Comparative genomic analysis of East Asian and non-Asian Helicobacter pylori strains identifies rapidly evolving genes. PloS one. 2013;8(1):e55120. [ Links ]
29. Kawai M, Furuta Y, Yahara K, et al. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiology. 2011;11:104. [ Links ]
30. Lara-Ramirez EE, Segura-Cabrera A, Guo X, Yu G, et al. New implications on genomic adaptation derived from the Helicobacter pylori genome comparison. PloS one. 2011;6(2):e17300. [ Links ]
31. Maldonado-Contreras A, Mane SP, Zhang XS, et al. Phylogeographic evidence of cognate recognition site patterns and transformation efficiency differences in H. pylori: theory of strain dominance. BMC Microbiology. 2013;13:211. [ Links ]
32. Sheh A, Chaturvedi R, Merrell DS, et al. Phylogeographic origin of Helicobacter pylori determines host-adaptive responses upon coculture with gastric epithelial cells. Infect Immun. 2013;81(7):2468-77. [ Links ]
33. Linz B, Windsor HM, Gajewski JP, et al. Helicobacter pylori genomic microevolution during naturally occurring transmission between adults. PloS one. 2013;8(12):e82187. [ Links ]
34. Akhter Y, Ahmed I, Devi SM, et al. The co-evolved Helicobacter pylori and gastric cancer: trinity of bacterial virulence, host susceptibility and lifestyle. Infect Agent Cancer. 2007;2(1):1-5. [ Links ]
35. Kodaman N, Sobota R, Mera R, et al. Disrupted human-pathogen co-evolution: a model for disease. Front Genet. 2014;5:290. [ Links ]
36. Loh JT, Shaffer CL, Piazuelo MB, et al. Analysis of cagA in Helicobacter pylori strains from Colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer Epidemiol Biomarkers Prev. 2011;20(10):2237-49. [ Links ]
37. Mane SP, Dominguez-Bello MG, Blaser MJ, et al. Host-interactive genes in Amerindian Helicobacter pylori diverge from their Old World homologs and mediate inflammatory responses. J Bacteriol. 2010;192(12):3078-92. [ Links ]
38. Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 2008;47(12):1077-83. [ Links ]
39. Lin D, Koskella B. Friend and foe: factors influencing the movement of the bacterium Helicobacter pylori along the parasitism-mutualism continuum. Evol Appl. 2015;8(1):9-22. [ Links ]
40. Chaturvedi R, de Sablet T, Asim M, et al. Increased Helicobacter pylori-associated gastric cancer risk in the Andean region of Colombia is mediated by spermine oxidase. Oncogene. 2015;34(26):3429-40. [ Links ]











 texto en
texto en