Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista colombiana de Gastroenterología
Print version ISSN 0120-9957
Rev Col Gastroenterol vol.33 no.2 Bogotá Apr./June 2018
https://doi.org/10.22516/25007440.168
Original articles
Genotyping of cagA and the intermediate region of vacA in strains of Helicobacter pylori isolated from Colombian adult patients and associations with gastric diseases
1Laboratorio de Diagnóstico Molecular y Bioinformática (LDMB). Universidad de los Andes. Bogotá D.C., Colombia.
2Max von Pettenkoffer Institute for Hygiene and Clinical Microbiology. LMU München. Department of Bacteriology. Múnich, Alemania.
3División de Gastroenterología y Endoscopía Digestiva. Fundación de Santa Fe de Bogotá. Bogotá D. C., Colombia
Objective:
This study characterizes the diversity of cagA and vacA virulence genes in Colombian patients to determine possible associations between them and the severity of endoscopic findings. It considers all four genotypes reported for the vacA gene (s, m and i).
Materials and methods:
Helicobacter pylori was detected in biopsies of 62 patients through culturing and by molecular methods. Genotypes of cagA and vacA (m/i/s) were determined by PCR and sequencing.
Results:
One hundred twenty four strains from 62 patients were isolated. Of these, 48.5% (n = 48) were vacA s2/m2/i2 - cagA (-) which were mostly found in patients with follicular gastritis; 32.3% (n = 32) were vacA s1/m1/i1-cagA (+) which were mostly found in patients with follicular gastritis, chronic gastritis and possible metaplasia. Significant associations were found between the presence of cagA and the vacA s1/m1/i1 genotype and the absence of cagA and the vacA s2/m2/i2 genotype (p <0.001). No significant association was found between the severity of endoscopic findings and the cagA-vacA status of the strains.
Conclusion:
We found a low prevalence of cagA (+) strains, the cagA-vacA status is not a predictor of risk in this population. Moreover, the presence of heterogenous infections without tropism suggests a need for biopsies from both the corpus and the antrum of the stomach in routine clinical practice.
Keywords: vacA subtypes; cagA; Helicobacter pylori; Colombia
Objetivo:
este estudio caracteriza la diversidad de los genes de virulencia cagA (gen asociado con la citotoxina A) y vacA (citotoxina vacuolizante) en pacientes colombianos para determinar posibles asociaciones entre estos 2 genes y la severidad de los hallazgos endoscópicos teniendo en cuenta todos los genotipos reportados para el gen vacA (s, m e i).
Materiales y métodos:
Helicobacter pylori fue detectado por cultivo y por métodos moleculares en biopsias de 62 pacientes. Los genotipos de cagA y vacA (m/i/s) se determinaron por reacción en cadena de la polimerasa (PCR) y secuenciación.
Resultados:
se aislaron 124 cepas de 62 pacientes; de estas, el 48,5% (n = 48) fueron vacA s2/m2/i2-cagA (-) presente en su mayoría en pacientes con gastritis folicular; mientras el 32,3% (n = 32) fueron vacA s1/m1/i1-cagA (+) presentes mayormente en pacientes con gastritis folicular, gastritis crónica y posible metaplasia. Se encontró una asociación significativa entre la presencia de cagA y el genotipo vacA s1/m1/i1 y la ausencia de cagA y el genotipo vacA s2/m2/i2 (p <0,001). No se encontró una asociación significativa entre la severidad de los hallazgos endoscópicos y el estatus cagA-vacA de las cepas.
Conclusión:
se encontró una baja prevalencia de cepas cagA (+), el estatus cagA-vacA no es un predictor de riesgo en la población estudiada y la presencia de infecciones heterogéneas sin tropismo sugieren la necesidad de tomar biopsias tanto del cuerpo como del antro del estómago en la práctica clínica rutinaria.
Palabras clave: Subtipos de vaca; cagA; Helicobacter pylori; Colombia
Introduction
Helicobacter pylori are spiral-shaped, microaerophilic gram-negative bacteria that infect more than 50% of the world’s population. 1 The infection has been associated with diseases such as gastritis, peptic ulcers, MALT (mucosa-associated lymphoid tissue) lymphoma and gastric adenocarcinoma. 2 H. pylori’s most important virulence factors are the cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA). 3 Depending on whether the cagA gene is present, strains are classified as positive (type I) which is more virulent due to the visible gastric damage induced, and negative (type II) which is less virulent and acts more like commensal bacteria than pathogens. 4
The VacA protein causes a series of alterations in human gastric cells that include formation of cytoplasmic vacuoles, 5,6 permeabilization of the plasma membrane, 6 mitochondrial secretion of cytochrome c, 7 mitochondrial fragmentation, 8 activation of mitogen-activated kinases. 9 and induction of autophagy. 6 It has been classified as an immunomodulatory protein. 6,10 The vacA gene has 3 polymorphic regions that are involved in the development of the disease: the signal region (s), the intermediate region (i), and the middle region (m). 2,3,6,11,12,13
The s region is involved in the efficiency of channel formation, and the m region affects tropism towards host cells. 3. Although the i region has only recently been described, it has been observed that the vacuolating activity of variant VacA i1 is stronger than that of subtype VacA i2. 3,6 Some analyses of the nucleotide and amino acid sequence of this gene have revealed polymorphisms grouped in clusters A, B and C. Of these, the amino acids in clusters B and C are responsible for the vacuolating activity of the protein. 11
To classify a strain as i1, i2 or i3, the amino acid sequence is compared with sequences of reference strains (60190 for i1 and Tx30a for i2). The i3 variant of vacA includes strains whose B cluster is similar to that of i1 and whose C cluster is similar to that of i2. 11 Different combinations of the 3 regions have been described: vacA s1/m1 strains have vacuolating activity and pose greater risk of gastric atrophy and adenocarcinoma than do the less virulent vacA s2/m2 strains. 11,12 vacA i1 strains have also been associated with gastric adenocarcinoma. 2,11,12
Recently, epidemiological studies have suggested that the interaction between cagA and vacA virulence factors, 6 factors of host susceptibility such as differences between men and women, 14,15,16 and environmental factors modulate development of the disease. 4,12
Several genotyping studies of H. pylori isolates from different Colombian populations have been published. Taking into account that the intermediate region can change the behavior of the toxin, this study complements data on genotyping of these two toxins including characterization of this region of vacA in isolates from a Colombian Andean population with gastric diseases of varying severity and evaluates correlations between genotype and the development of gastric disease.
Materials and methods
Patients and Study Population
We included 62 patients from Bogotá, Colombia who were over the age of 18 years, who were infected with H. pylori, and who had gastric symptoms and indications for endoscopy. Patients who had ingested proton pump inhibitors (PPI) within the 15 days prior to the procedure, antacids within the 12 hours prior to the procedure, or antibiotics in month prior to the procedure were excluded. Also excluded were patients suffering from cardiovascular and respiratory diseases, cancer patients who had undergone radiation therapy or chemotherapy in the six months prior to the procedure, and patients with coagulopathies or amyloidosis.
Strains and DNA Purification
Biopsies were macerated and a 10-1 dilution was seeded on GC agar (Oxoid, Germany) supplemented with cholesterol (1X, Gibco) and DENT (Oxoid, Germany). 17 They were incubated at 37° C in a controlled atmosphere with 10% carbon dioxide (CO2) for four to ten days. The Quick-gDNA Miniprep kit (Zymo Research, California, United States) was used to extract DNA in accordance with the manufacturer’s instructions. The strains were selected by culture and confirmed as H. pylori by polymerase chain reaction (PCR) of the 23S gene with the HPY-S and HPY-A primers previously reported. 18
Detection of cagA
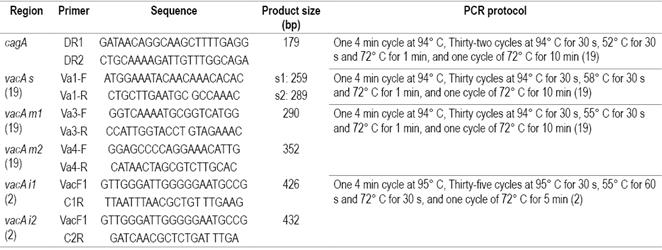
PCR was performed with the primers designed in this study and the previously reported protocol as modified and described in Table 1. 19 The PCR had a final volume of 25 μL with GoTaq Green Master Mix 1X (Promega, Wisconsin, United States) and 0.25 mM of each primer. Visualization was performed on 1.5% agarose gels.
vacA Genotyping
The 3 polymorphic regions of the vacA gene were characterized by means of PCR with a final volume of 25 μL containing GoTaq Green Master Mix 1X (Promega, Wisconsin, United States) and 0.25 mM of the corresponding primers for each region according to the previously reported protocols as modified in this study (Table 1).
For the visualization of the products, 2% agarose gels were used for the m and i regions and 10% polyacrylamide was used for the s region. (29: 1) Polyacrylamide gels were stained with a 3X GelRed solution (15 μL of Gelred 10,000X and 5 mL of sodium chloride [NaCl] 1M) for 1 hour under constant stirring at room temperature.
Samples that tested positive for both subtypes (vacA i1 and i2) were sequenced (Macrogen, Seoul, Korea). Sequence analysis was performed with CLC Genomics Workbench 10, and amino acid sequences were compared with reference sequences 60190 (GenBank No. U05676) and Tx30a (GenBank No. U29401). Strains were defined as i3 when the B cluster was similar to that of reference strain i1 and the C cluster was similar to reference strain i2.11
Statistical Analysis
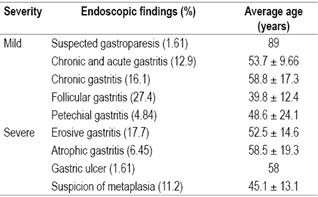
Fisher’s exact test was used to determine possible associations between the cagA status and the vacA genotype of the strains, and a generalized linear model was used to determine which variables had significant associations. The same statistical analysis was performed for polymorphic regions of the vacA gene and cagA status and between and among cagA status, vacA gene subtypes and severity of endoscopic findings. A chi squared (χ2) test was used to determine associations between the patient’s sex and cagA status, and the patient’s sex and severity of endoscopic findings. Endoscopic findings were classified as either mild gastric disease and severe gastric disease (Table 2). R software 20 was used for analysis and a p value <0.05 was considered significant.
Results
Patients and Isolation of H. pylori
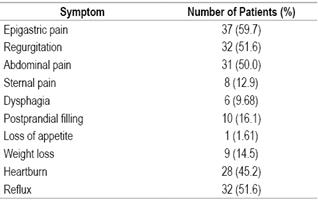
We analyzed 124 H. pylori strains from 62 patients who had a variety of symptoms (Table 3). Their average age was 50.2 ± 16.2 years, and the male-female ratio was 1.48/1.
Genotyping of cagA and vacA
PCR with corresponding primers established that 52 (48.1%) of the 124 strains analyzed were positive for cagA. No significant associations were found between the severity of endoscopic findings and the presence of cagA (χ2, p> 0.05) (Table 4). There were also no significant associations between the sex of the patients and the presence of cagA (χ2, p> 0.05), between the severity of the endoscopic findings and sex (Fisher’s exact test, p> 0.05), or between the severity of the endoscopic findings and the presence of cagA (χ2, p> 0.05) (Table 4).
Table 4 Association between severity of endoscopic findings, cagA status and polymorphic regions of the vacA gene*
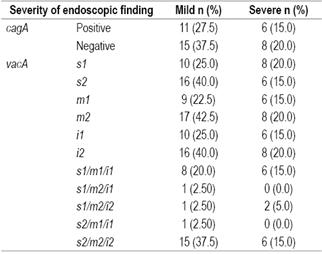
* No significant relationships were found between the severity of endoscopic findings and cagA or vacA status. Patients who had different genotypes in the gastric antrum and corpus were excluded. Fisher’s exact test and a generalized linear binomial model (p> 0.05) were used.
In the case of vacA, 16 strains were discarded (n = 8 patients) because it was not possible to determine the genotype for the s region in three cases (37.5%), the m region in three cases (37.5%), the s region in one case (12.5%) and for all three regions in one other case (12.5%). Of the remaining 108 strains, fifty strains were s1, fifty-eight were s2, forty-one were m1, and sixty-seven were m2. Thirty-four strains were i1 and 55 i2. Nineteen of the samples sequenced (17.6%) showed amplification for both vacA i1 and vacA i2 subtypes (GenBank No. MF457450 to MF457477). Of these, four were classified as i1, three as i2, three as i3 and nine as i1i2. The last group was eliminated in subsequent analyses. No significant associations were found the severity of the endoscopic findings and either vacA subtypes or the full vacA genotype (χ2, p> 0.05) (Table 4).
Twenty-eight strains (22.6%) were discarded from the evaluation of the relationship between the two genes because they had different genotypes of cagA and vacA between antrum and corpus. No significant tropism or correlation was found between the genotype and location within the stomach of these strains. However, a significant association was found between cagA status and the vacA gene subtypes looked at separately (Fisher’s exact test, p <0.05) (Figure 1) and combined as follows: s1/m1/i1-cagA positive (32.3%), s1/m2/i1-cagA positive (2.02%) and s1/m2/i2 (7.07%). Of these 71.4% were cagA positive, 2.02% were s1/m1/i3-cagA positive, 1.01% were s1/m2/i3-cagA positive, 2.02% were s2/m1/i1-cagA positive, and 48.5% were s2/m2/i2-cagA negative (Fisher’s exact test, p <0.001, generalized linear model binomial p <0.05). In addition, all vacA i3 strains were cagA positive.
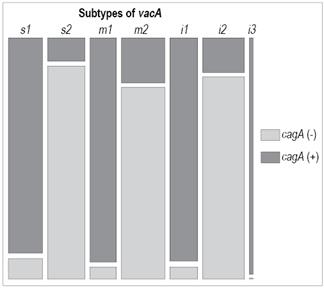
Figure 1 Association between cagA status and vacA subtypes. There is a significant association between cagA status and vacA subtypes. Patients with cagA status or vacA genotypes that differed between the antrum and corpus were excluded. Fisher’s exact test and a generalized linear binomial model (p <0.001) were used.
Discussion
This study characterizes the diversity of virulence genes cagA and vacA in Colombian patients to determine possible associations between these two genes and the severity of the endoscopic findings. It considers all three genotypes reported for the vacA gene: s, m and i. The combination of genetic variability, host factors and environmental factors plays a fundamental role in the development of pathologies during H. pylori infections. 3,4,6,12,21,22,23. It has been reported that patients infected with vacA s1/m1/i1-cagA (+) strains are at greater risk of developing gastric carcinomas than are patients infected with the less virulent vacA s2/m2/i2-cagA (-). 2,4,11,23,13,19,24
In the Colombian population studied, a prevalence of 48.1% was found for infections with cagA positive strains. This is lower than previously reported in studies conducted in Bogotá, Colombia which found a prevalence of 73%, 24 but it is similar to that reported in patients from Tolima, Colombia for whom prevalence of cagA positive strains was 43%. 25 The prevalence reported here also differs from those found in other regions of the world such as Senegal (73.3%), 26 the Middle East (100%), the United States (80%), and Western Europe and Latin America (60% to 70%). 11 This is probably due to the fact that the selected population was of medium and high socioeconomic class since it has been seen that the prevalence of the infection is higher in lower socioeconomic levels with poor sanitary conditions. 27,28
This study also shows that 22.6% of the patients were infected with two different strains of either or both cagA and vacA in the gastric antrum and corpus but without any tropism. It is possible that these patients have been infected on more than one occasion by different strains of H. pylori because the prevalence of multiple infections seems to be higher in places like Colombia where the risk of H. pylori infection is high. 29,30 The presence of these types of infections reaffirms the need to routinely take biopsies from both the corpus and antrum.
The relationship between the presence of cagA and the vacA s1 genotype has been described by several authors. 31,32 In this study, 68.1% of positive cagA strains were vacA s1/m1/i1 and only 6.4% were vacA s2/m2/i2. Thus, the vacA s1/m1/i1 genotype has a significant association with the presence of the cagA gene. In the case of vacA s1/m2 strains, it has been observed that whether the prevalence of type i1 is greater that the prevalence of i2 strains and vice versa depends on the geographical region. 2,26 A low prevalence of the vacA s2/m1 combination was found in this study. This agrees with previous findings which suggests that the low prevalence of this genotype may be explained by negative selection of this subtype because it is unfavorable for microorganisms. 26
In addition, this study describes intermediate region of the vacA gene strains in strains circulating in this Colombian population. Almost all of the vacA s1/m1 strains (94.4%) were i1 and almost all the vacA s2/m2 strains were i2. This agrees with what was reported in the first description of the intermediate region. 2 The majority of strains analyzed were vacA i2, and only three patients were found to be infected by vacA i3 strains defined as those with cluster B similar to i1 and cluster C similar to type i2. (11) The i3 variant could reflect a recombination process between type i1 and i2 strains. 6,11 The clinical or pathological importance of this variant remains unknown, and our study found no significant associations with endoscopic findings. However, all vacA i3 strains were cagA positive. A larger sample of patients infected with i3 strains is needed to gain a better understanding of the nature of this genotype and the development of the disease in these patients.
In contrast to the significant associations between the vacA s1/m1/i1-cagA (+) genotype and some severe digestive diseases such as atrophic gastritis, ulcers and gastric adenocarcinoma observed in other studies, 33 no significant associations were found between the cagA-vacA status of the strains and the severity of the endoscopic findings in this study despite the fact that most of the strains isolated were vacA s2/m2/i2-cagA (-) from patients who mostly presented follicular gastritis (Figure 2). This could be the result of the small sample sizes of each of the categories of severity of endoscopic findings (mild or severe) and the high prevalence of type II strains in the patients. For this reason, the vacA genotype cannot be used to identify high-risk patients in the study population.
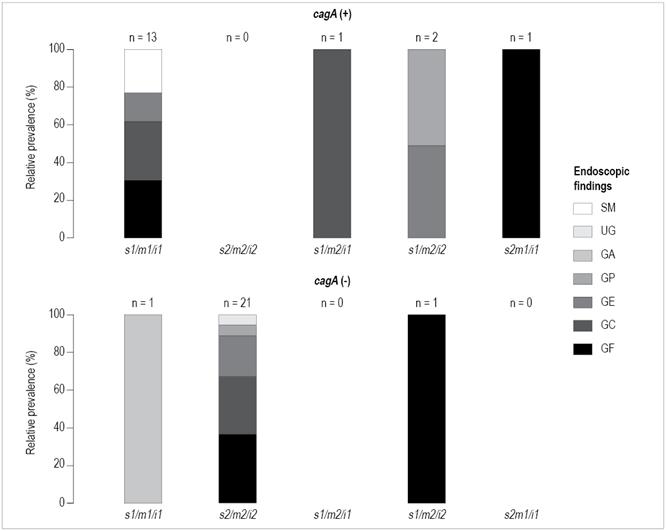
Figure 2 Relative prevalence of endoscopic findings according to the genotype of H. pylori strains isolated in these patients. 52.5% of patients were infected with H. pylori vacA s2/m2/i2-cagA negative strains which were mostly related to follicular gastritis. Patients who had different cagA or vacA genotypes in the antrum and corpus were excluded. Suspicion of metaplasia (SM), gastric ulcer (UG), atrophic gastritis (GA), petechial gastritis (GP), erosive gastritis (GE), chronic gastritis (GC) and follicular gastritis (GF).
In conclusion, this study provides new information on the prevalence of cagA and vacA virulence factors, and their association with endoscopic findings. Studies conducted on the west coast and in the southern Colombian Andes differ from the findings of this investigation highlighting the need to increase the number of studies that compare different regions and Colombian populations. 34,35 This is especially true since the data published in Colombia show great variations in the presence of genetic markers of pathogenicity and their correlations with the severity of gastric pathologies.
On the other hand, recent studies have indicated that H. pylori strains in the Colombian population originating in the European line (HpEurope) have evolved independently. 36 It is likely that molecular mechanisms typically associated with severe gastric diseases associated H. pylori behave differently in the Colombian population thanks to the different ancestries present. 37 If the bacterium-host relationship is considered, it is possible that the lack of correlations between cagA and vacA and the pathology of the infection relative to the findings of other studies could be thoroughly investigated only if the ancestral distributions in the various regions of Colombia.
Acknowledgements
The authors would like to thank Sebastián Posada MD, Laura Camila Zuluaga MD, Daniela Campaz, MD and Manuel Arrieta MD for their help in the collection of samples and management of the database. We would also like to thank Verónica Arévalo Jaimes for her help with molecular processing and identification of strains by means of PCR of the 23S region. We thank Professor Adolfo Amézquita and Cindy Ulloa for their support in statistical analysis.
REFERENCES
1. Peleteiro B, Bastos A, Ferro A, et al. Prevalence of Helicobacter pylori infection worldwide: a systematic review of studies with national coverage. Dig Dis Sci. 2014;59(8):1698-709. doi: 10.1007/s10620-014-3063-0. [ Links ]
2. Rhead JL, Letley DP, Mohammadi M, et al. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133(3):926-36. doi: 10.1053/j.gastro.2007.06.056. [ Links ]
3. Jones KR, Jang S, Chang JY, et al. Polymorphisms in the intermediate region of VacA impact Helicobacter pylori-induced disease development. J Clin Microbiol. 2011;49(1):101-10. doi: 10.1128/JCM.01782-10. [ Links ]
4. García CA, Barra TR, Delgado SC, et al. Genotipificación de aislados clínicos de Helicobacter pylori en base a genes asociados a virulencia cagA, vacA y babA2: primer aislamiento de una cepa babA2 positiva en pacientes chilenos. Rev Médica Chile. 2006;134(8):981-8. doi: 10.4067/S0034-98872006000800006. [ Links ]
5. Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267(15):10570-5. [ Links ]
6. Kim I-J, Blanke SR. Remodeling the host environment: modulation of the gastric epithelium by the Helicobacter pylori vacuolating toxin (VacA). Front Cell Infect Microbiol. 2012;2:37. doi: 10.3389/fcimb.2012.00037. [ Links ]
7. Foo JH, Culvenor JG, Ferrero RL, et al. Both the p33 and p55 Subunits of the Helicobacter pylori VacA Toxin Are Targeted to Mammalian Mitochondria. J Mol Biol. 2010;401(5):792-8. doi: 10.1016/j.jmb.2010.06.065. [ Links ]
8. Jain P, Luo Z-Q, Blanke SR. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc Natl Acad Sci U S A. 2011;108(38):16032-7. doi: 10.1073/pnas.1105175108. [ Links ]
9. Nakayama M, Kimura M, Wada A, et al. Helicobacter pylori VacA Activates the p38/Activating Transcription Factor 2-mediated Signal Pathway in AZ-521 Cells. J Biol Chem. 2004;279(8):7024-8. doi: 10.1074/jbc.M308898200. [ Links ]
10. Boncristiano M, Paccani SR, Barone S, et al. The Helicobacter pylori vacuolating toxin inhibits T cell activation by two independent mechanisms. J Exp Med. 2003;198(12):1887-97. doi: 10.1084/jem.20030621. [ Links ]
11. Chung C, Olivares A, Torres E, et al. Diversity of vacA intermediate region among Helicobacter pylori strains from several regions of the world. J Clin Microbiol . 2010;48(3):690-6. doi: 10.1128/JCM.01815-09. [ Links ]
12. Ferreira RM, Machado JC, Letley D, et al. A novel method for genotyping the Helicobacter pylori vacA Intermediate region directly in gastric biopsy specimens. J Clin Microbiol . 2012;50(12):3983-9. doi: 10.1128/JCM.02087-12. [ Links ]
13. González-Rivera C, Algood HMS, Radin JN, et al. The intermediate region of Helicobacter pylori vacA is a determinant of toxin potency in a jurkat t cell assay. Infect Immun. 2012;80(8):2578-88. doi: 10.1128/IAI.00052-12. [ Links ]
14. Agah S, Khedmat H, Ghamar-Chehred ME, et al. Female gender and Helicobacter pylori infection, the most important predisposition factors in a cohort of gastric cancer: a longitudinal study. Casp J Intern Med. 2016;7(2):136-41. [ Links ]
15. Naja F, Kreiger N, Sullivan T. Helicobacter pylori infection in Ontario: Prevalence and risk factors. Can J Gastroenterol. 2007;21(8):501-6. doi: 10.1155/2007/462804. [ Links ]
16. Zhu Y, Zhou X, Wu J, et al. Risk factors and prevalence of Helicobacter pylori infection in persistent high incidence area of gastric carcinoma in Yangzhong City. Gastroenterol Res Pract. 2014;2014:e481365. [ Links ]
17. Jiménez-Soto LF, Rohrer S, Jain U, et al. Effects of cholesterol on Helicobacter pylori growth and virulence properties in vitro. Helicobacter. 2012;17(2):133-9. doi: 10.1111/j.1523-5378.2011.00926.x. [ Links ]
18. Ménard A, Santos A, Mégraud F, et al. PCR-restriction fragment length polymorphism can also detect point mutation A2142C in the 23S rRNA gene, associated with Helicobacter pylori resistance to clarithromycin. Antimicrob Agents Chemother. 2002;46(4):1156-7. doi: 10.1128/AAC.46.4.1156-1157.2002. [ Links ]
19. Secka O, Antonio M, Berg DE, et al. Mixed infection with cagA positive and cagA negative strains of Helicobacter pylori lowers disease burden in The Gambia. PLoS ONE. 2011;6(11):e27954. doi: 10.1371/journal.pone.0027954. [ Links ]
20. R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing ENT#091;internetENT#093; 2008 ENT#091;acceso el 24 de noviembre de 2016ENT#093;. Disponible en: Disponible en: http://www.R-project.org . [ Links ]
21. Argent RH, Thomas RJ, Letley DP, et al. Functional association between the Helicobacter pylori virulence factors vacA and cagA. J Med Microbiol. 2008;57(2):145-50. doi: 10.1099/jmm.0.47465-0. [ Links ]
22. Jang S, Jones KR, Olsen CH, et al. Epidemiological link between gastric disease and polymorphisms in vacA and cagA. J Clin Microbiol . 2010;48(2):559-67. doi: 10.1128/JCM.01501-09. [ Links ]
23. Yamazaki S, Yamakawa A, Okuda T, et al. Distinct Diversity of vacA, cagA, and cagE Genes of Helicobacter pylori Associated with Peptic Ulcer in Japan. J Clin Microbiol . 2005;43(8):3906-16. doi: 10.1128/JCM.43.8.3906-3916.2005. [ Links ]
24. Quiroga AJ, Cittely DM, Bravo MM. Frecuencia de los genotipos babA2, oipA y cagE de Helicobacter pylori en pacientes colombianos con enfermedades gastroduodenales. Biomédica. 2005;25:325-34. doi: 10.7705/biomedica.v25i3.1357. [ Links ]
25. Molina Delgado A, Jaramillo Henao C, Delgado Perafán M, et al. Detección y genotipificación de Helicobacter pylori sobre la base de los genes ADNr 16S y el gen asociado a citotoxina (cagA) y posible asociación con enfermedades gastrointestinales. Revista Cubana de Medicina Tropical. 2008;60:105-10. [ Links ]
26. Basso D, Zambon C-F, Letley DP, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135(1):91-9. doi: 10.1053/j.gastro.2008.03.041. [ Links ]
27. Parente JML, da Silva BB, Palha-Dias MPS, et al. Helicobacter pylori infection in children of low and high socioeconomic status in northeastern Brazil. Am J Trop Med Hyg. 2006;75(3):509-12. [ Links ]
28. Ramírez Ramos A, Chinga Alayo E, Mendoza Requena D, et al. Variación de la prevalencia del H. pylori en el Perú período (1985-2002), en una población de nivel socioeconómico medio y alto. Rev Gastroenterol Perú. 2003;23(2):92-8. [ Links ]
29. Mendoza-Elizalde S, Cortés-Márquez AC, Giono-Cerezo S, et al. Analysis of the genotypic diversity of strains of Helicobacter pylori isolated from pediatric patients in Mexico. Infect Genet Evol. 2015;29:68-74. doi: 10.1016/j.meegid.2014.11.002. [ Links ]
30. Pinto-Ribeiro I, Ferreira RM, Batalha S, et al. Helicobacter pylori vacA genotypes in chronic gastritis and gastric carcinoma patients from Macau, China. Toxins (Basel). 2016;8(5). pii: E142. doi: 10.3390/toxins8050142. [ Links ]
31. Atherton JC, Cao P, Peek RM Jr, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):1-7. doi: 10.1074/jbc.270.30.17771. [ Links ]
32. Nagiyev T, Yula E, Abayli B, et al. Prevalence and genotypes of Helicobacter pylori in gastric biopsy specimens from patients with gastroduodenal pathologies in the Cukurova Region of Turkey. J Clin Microbiol . 2009;47(12):4150-3. doi: 10.1128/JCM.00605-09. [ Links ]
33. Breurec S, Michel R, Seck A, et al. Clinical relevance of cagA and vacA gene polymorphisms in Helicobacter pylori isolates from Senegalese patients. Clin Microbiol Infect. 2011;18(2):153-9. doi: 10.1111/j.1469-0691.2011.03524.x. [ Links ]
34. Bravo MM, Trujillo E, Quiroga A, et al. Genotipificación de Helicobacter pylori en individuos de dos regiones de Colombia con riesgo de cáncer gástrico opuesto. Rev Colomb Cancerol. 2013;17(4):178-9. [ Links ]
35. Nogueira C, Figueiredo C, Carneiro F, et al. Helicobacter pylori genotypes may determine gastric histopathology. Am J Pathol. 2001;158(2):647-54. doi: 10.1016/S0002-9440(10)64006-0. [ Links ]
36. Muñoz-Ramírez ZY, Mendez-Tenorio A, Kato I, et al. Whole genome sequence and phylogenetic analysis show Helicobacter pylori strains from Latin America have followed a unique evolution pathway. Front Cell Infect Microbiol. 2017;7:50. doi: 10.3389/fcimb.2017.00050. [ Links ]
37. Ossa H, Aquino J, Pereira R, et al. Outlining the ancestry landscape of Colombian admixed populations. PLoS One. 2016;11(10):e0164414. doi: 10.1371/journal.pone.0164414. [ Links ]
Financing This study was supported by the Faculty of Sciences of the Universidad de los Andes through the project entitled “Factors of virulence cagA and vacA of strains of H. pylori and its relationship with pathology in symptomatic Colombian adult patients” code P16.160322.001/01-BIO01 (according to a decision of the Research Projects Committee on 12/11/2015) and by DFG research grant (JI 221/1-1) for LFJ-S.
Received: September 28, 2017; Accepted: April 13, 2018











 text in
text in