Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Agronomía Colombiana
Print version ISSN 0120-9965
Agron. colomb. vol.29 no.1 Bogotá Jan./Apr. 2011
Gene silencing and applications for functional gene validation: The case of Geminiviruses
Silenciamiento génico y aplicaciones para la validación funcional de genes: el caso de los Geminivirus
Simón Pedro Cortés1 and Camilo Ernesto López2,3
1 Doctoral Program in Science-Biology, Biodiversity and Conservation line, Universidad Nacional de Colombia. Bogota (Colombia).
2 Department of Biology, Faculty of Sciences, Universidad Nacional de Colombia. Bogota (Colombia).
3 Corresponding author: celopezc@unal.edu.co
Received for publication: 22 January, 2010. Accepted for publication: 2 February, 2011.
ABSTRACT
Plants are able to recognize and degrade double-strand RNA molecules employing the mechanism of post-transcriptional gene silencing (PTGS). PTGS is both a vegetal defense strategy against viral infections and a conserved eukaryotic mechanism for regulate endogenous gene expression. The VIGS methodology (Virus Induced Gene Silencing) uses this mechanism to selectively silence genes employing viral vectors, which contain the target gene, becoming a tool for functional gene validation. Geminiviridae family, with the Begomovirus, Curtovirus, Mastrevirus and Topocuvirus genera, encompass viruses of circular, single-strand DNA packed in icosahedric geminated particles, and some of them had been evolved to infect particular plant species. Classification of these genera is based in the genome organization, the type of host plants and the insect vector that transmits the virus. Geminiviruses are able to induce gene silencing (GS) and therefore they have been used to develop VIGS-based methodologies for functional gene silencing. This review describes the molecular mechanism of gene silencing, with emphasis in gene silencing induced by Geminiviruses and the applications for a staple crop as cassava.
Key words: gene silencing, functional genomics, Geminivirus, Manihot esculenta.
RESUMEN
Las plantas reconocen y degradan moléculas de ARN de doble cadena a través del mecanismo de silenciamiento génico post-transcripcional (PTGS). Se trata de una estrategia de defensa vegetal contra infecciones virales y, simultáneamente, de un mecanismo conservado en los eucariotas para regular la expresión endógena de genes. La metodología VIGS (Virus Induced Gene Silencing) aprovecha el PTGS para silenciar genes selectivamente empleando vectores virales que contienen el gen blanco, constituyéndose en una herramienta para la validación funcional de genes. La familia Geminiviridae, con los géneros Begomovirus, Curtovirus, Mastrevirus y Topocuvirus comprende virus con ADN de cadena circular sencilla empacado en partículas icosahédricas geminadas, algunos de los cuales han evolucionado para infectar especies particulares de plantas. La clasificación de estos géneros se basa en la organización del genoma, el rango de plantas hospederas y el insecto vector que los transmite. Los Geminivirus inducen silenciamiento génico (SG) y han sido empleados como herramientas de VIGS para la validación funcional de genes. Esta revisión describe la base molecular del mecanismo de silenciamiento génico con énfasis en el silenciamiento inducido por Geminivirus y las aplicaciones a un cultivo de seguridad alimentaria como la yuca.
Palabras clave: silenciamiento génico, genómica funcional, Geminivirus, Manihot esculenta.
Introduction
Viral diseases represent great limitations in crop production of agronomic interest. In response to viral infection, plants have developed different kinds of immunities, one of which involves gene silencing (GS), which produces the degradation of the double-strand RNA molecules of viral origin produced in the process of virus replication. This phenomenon has been used to implement strategies for the functional validation of genes, inhibiting gene expression and evaluating the resulting phenotype. This is how the biotechnological exploitation of plant-virus interactions has permitted elucidating the function of multiple genes. This review conducts a description of the GS mechanism with emphasis to that produced by Geminiviruses and its application for the functional validation of genes in plants.
Types of silencing
Gene silencing controls viral replication within the host cells and avoids the invasion of characteristic DNA fragments like transposons and regulates gene endogenous expression (Chapman and Carrington, 2007). GS is highly conserved evolutionarily and involves the degradation of RNA molecules (Chen and Rajewsky, 2007). It was initially reported in plants, where it was denominated as Post Transcriptional Gene Silencing (PTGS) (Napoli et al., 1990), but it was subsequently described in fungus (quelling) (Cogoni and Macino, 1997) and recently in animals, where it is known as RNA interference (RNAi) (Fire et al., 1998).
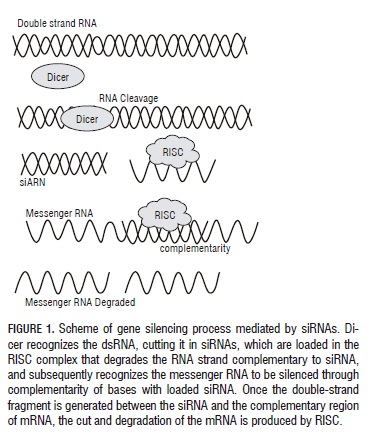
GS consists of the junction between a short RNA sequence of approximately 20-30 nucleotides (nts) with an RNA target sequence for its degradation (Fire et al., 1998). There are several paths activating the degradation mechanism of RNA molecules, which is related to the evolutionary histories of each of the different organisms and the functions GS must fulfill (Voinnet, 2009). GS starts with the presence of a double-strand RNA (dsRNA) recognized by the Dicer enzyme, which has RNAase lll activity and, specifically cuts dsRNAs producing duplex RNAs of 21 to 24 nts called siRNA (small interfering RNAs). Dicer is evolutionarily conserved in diverse organisms (Bernstein et al., 2001). The siRNAs generated by Dicer are recruited by the multienzyme RISC complex (RNA-Induced Silencing Complex), which contains Argonaut (AGO) (Hammond et al., 2001). Within RISC, one of the siRNA strands remains joined to the complex and the complementary strand is hydrolyzed, possibly by AGO (Hutvagner and Simard, 2008). When RISC has been charged with siRNA, it begins to recognize messenger RNA sequences (mRNA) complementary to siRNA, cutting them off or repressing their translation (Baulcombe, 2005) (Fig. 1). Silencing may extend due to a RNA-dependent RNA polymerase (RDRP), which amplifies the signal producing additional siRNAs migrating from the infection site to remote cells (Voinnet, 2008).
The origin of the dsRNAs defines the different GS mechanisms in the organisms. RNA silencing may occur at the transcriptional level (Transcriptional Gene Silencing, TGS), as well as at the post-transcriptional (PTGS) level (Brodersen and Voinnet, 2006). In Drosophila germ cells, a type of small (between 24-30 nts), noncoding RNA has been described, denominated piwiRNA, which originates from a single-strand RNA precursor without needing the Dicer. The Piwi complex would join the transcripts emerging from the transposable elements (TE), recruiting chromatin remodeling or assembly factors and activating the transcription (Klattenhoff and Theurkauf, 2008).
TGS involves the methylation of promoter regions reducing or inactivating the genetic expression (Bao et al., 2004). This silencing is related to controlling the TE invasion in the genome (Obbard et al., 2009). Methylation-based GS is begun by small RNAs (sRNAs) of 24 nts from transposons (Mosher et al., 2008). The sRNAs induce epigenetic silencing by generating RNA-directed DNA methylation (RdRM) (Chan et al., 2005)
PTGS is mediated by microRNAs (miRNAs). The miRNAs are a fundamental part for the regulation of the expression of endogenous genes in plants and animals and participate in multiple processes and responses to biotic and abiotic stresses (Brodersen and Voinnet, 2009). miRNAs are transcribed by RNA polymerase II, forming the primary miRNA (Lee et al., 2002). These primary miRNAs are processed in the nucleus by Drosha-type proteins making up miRNAs precursors, which are exported to cytoplasm and processed by Dicer, forming mature miRNAs (Chapman and Carrington, 2007). As with siRNA from the virus (see below), miRNAs are recruited by RISC where one of the RNA strands (denominated passenger RNA*) is lost. The miRNA:RISC complex interacts with the complementary mRNA molecule producing its cut and degradation (Voinnet, 2009).
The first PTGS case was discovered in plants as a defense mechanism against viral infections (Hamilton and Baulcombe, 1999). Many virus infecting plants are made up of single-strand RNA, which in order to replicate go through an intermediary stage where double-strand molecules are produced, which are recognized by Dicer and cut into siRNAs (Voinnet, 2009). These siRNAs are incorporated to RISC, where a strand of the siRNA is cut and degraded.
The single-strand siRNA remaining in the RISC, being complementary to the viral RNA sequence, will make up a duplex where the viral RNA is digested, generating resistance to the infection (Baulcombe, 2005). In various eukaryotes, there is RDRP, which permit producing more siRNAs, amplifying the signal and making it systemic (Ding and Voinnet, 2007).
Suppressors
The interaction between plants and virus can be seen as an "arms race" (Waterhouse and Fusaro, 2006). In this sense, viruses have generated mechanisms to block the GS and in consequence the resistance of the host (Deleris et al., 2006) through silencing suppressors, initially characterized as viral pathogenicity factors (Voinnet et al., 1999). GS suppressor proteins are not similar at the sequence level, but act at different levels within the same GS route, suggesting they have originated via convergent evolution processes (Soosaar et al., 2005). The first suppressor identified was HC-Pro in Potyvirus (Pruss et al., 1997). HC-Pro acts as a suppressor joining Dicer, interfering with its activity and trapping the siRNAs (Voinnet, 2005). Other suppressors acting similarly are P19 from the Tombusvirus (Tombusviridae) and P21 from BYV (Beet yellow virus) (Lakatos et al., 2006), P38 from TCV (Turnip crinkle virus), and 2b from CMV (Cucumber mosaic virus) (Deleris et al., 2006). Recently, P0 was characterized from BWYV (Beet western yellow virus), which presents an F-box motive implicated in protein degradation and interacts with AGO degrading it and blocking GS (Bortolamiol et al., 2007).
Gene silencing and functional genomics
The most widely used mechanism to determine gene function is the generation of mutants of genes of interest, causing the loss of function or expression through mutations in the promoters (Page and Grossniklaus, 2002). The collection of mutants generated is evaluated to identify the resulting phenotype. The way of applying this methodology involves the insertion of DNA sequences (transposons or T-DNA elements in plants) (Page and Grossniklaus, 2002), homologous recombination (bacteria and animals) (Kile and Hilton, 2005; Shuman and Silhavy, 2003), treatment with mutagenic agents like sodium azide (Rines, 1985), ethyl methanesulfonate (EMS) (Katavic et al., 1995), or gamma radiation exposure (Vizir et al., 1994) to just name the most important. Identifying the GS mechanism in plants and animals generated new methodological strategies that have permitted guided silencing of target genes for their functional validation (Boutros and Ahringer, 2008).
In Drosphila melanogaster and Caenorhabditis elegans, silencing has been implemented by RNAi (Dorsett and Tuschl, 2004). Initially long dsRNA were employed, which were cleaved in intra-cellular manner by Dicer in functional siRNAs (Fire et al., 1998; Hammond et al., 2000). Nevertheless, these long dsRNAs are not very efficient in activating the silencing in many mammal cells (Grimm, 2004). Alternatively, cells are transfected with synthetic siRNAs or cloned in plasmids (Jackson et al., 2006; Berns et al., 2004). Thus using for example, siRNA libraries, functional genomics experiments have been conducted to determine the phenotypic effect of inactivating the genes annotated through complete genome sequencing strategies (Berns et al., 2004; Boutros et al., 2004; Paddison et al., 2004). There are siRNA libraries available in public and private research centers (Boutros and Ahringer, 2008).
For C. elegans, the ingestion of bacteria containing plasmids with siRNAs permits introducing these into the nematode cell cytoplasm, inducing silencing of the endogenous target gene (Jorgensen and Mango, 2002), while in Drosophila or cell cultures it is necessary to transfect to incorporate siRNAs into the cytoplasm (Boutros and Ahringer, 2008).
In plants, the Virus-Induced Gene Silencing (VIGS) strategy was developed in which a gene or part of such is introduced into a viral vector in antisense direction (Eamens et al., 2008), is inoculated in plants and a couple of weeks later the silencing of the endogenous gene occurs, by producing siRNAs generated by Dicer from dsARN produced during viral replication, and which includes the production of dsRNAs complementary to the target gene to be silenced. The VIGS system has been widely used mainly in plants from the Solanaceae family while in others it has been difficult to implement. It can be explained considering the difficulty finding a balance between the replication capacity of the viral construct and the plant's relative resistance against infection. To avoid a phenotype product of the virus instead of the target gene to be silenced is troublesome (Purkayastha and Dasgupta, 2009). Alternatively, the strategy known as RNAi has been developed, which consists of introducing, via genetic transformation, a sense-antisense construction of the target gene to be silenced. This way, the transcription of the "transgen" in the plant cell will produce dsRNAs, which will be recognized by Dicer and siRNAs are generated complementary to the endogenous gene to silence (Wesley et al., 2001). This strategy takes up more time given that transgenic lines must be generated with the silencing construct; this is a rather inefficient process in plants recalcitrant to genetic transformation (Purkayastha and Dasgupta, 2009).
Geminiviruses are part of the game
Although most viruses infecting plants have RNA genomes, there are exceptions like viruses from the Caulimoviridae, Geminiviridae, and Circoviridae families; curiously, in prokaryotes, invertebrates, and vertebrates, most viral infections are caused by DNA viruses (Rojas et al., 2005). Caulimoviruses are formed by dsDNA, which is replicated via an RNA intermediary by reverse transcription (Schoelz, 2008). Geminiviruses and Nanoviruses have single-strand DNA or ssDNA, replicating via an intermediary dsDNA molecule through the rolling circle mechanism (Hanley- Bowdoin et al., 2000).
Since the beginning of 20th century, the first plant infections caused by Geminiviruses were recognized, although they were much later associated to vector insects. Isolation and characterization of Geminiviruses was undertaken in the 1970s (Mumford, 1974). Geminiviral diseases affect production of crops like corn, bean, cassava and tomato (Fargette et al., 2004; Mansoor et al., 2006), to name just some cases.
Geminiviruses have a genome composed of one (monopartite) or two (bipartite) circular ssDNA molecules whose sizes range between 2.5 and 3.0 kb. These viruses use a mechanism for bidirectional transcription and genetic overlap that maximizes the use of their genome (Rojas et al., 2005). Structurally, they present twin icosahedral virions, which gives them their name (from the Latin geminis: twin) (Matthews and Hull, 2002). Geminiviruses present the REP protein (replication-associated protein), essential for replication, and whose genomic position, sequence and function is conserved in all the Geminiviruses (Matthews and Hull, 2002). Geminiviruses are grouped in the Geminiviridae family, made up of four genus according to their genomic arrangement, host range, and vector specificity: Geminiviruses with monopartite genomes and transmitted by insects from the Cicadellidae family to monocotyledon plants belong to the Mastrevirus genus. Geminivirus with the characteristics of Mastrevirus but transmitted to dycotyledon plants are Curtovirus. The Topocuvirus genus is monopartite and infects dicotyledon plants through insects from the Membracidae family. The Begomovirus genus is one of the most diverse Geminiviruses; it is transmitted exclusively by the Bemisia tabaci whitefly complex to dycotyledon plants and contains mono and bipartite members (Vanitharani et al., 2005).
One of the evolutionary advantages permitting Geminiviruses (like DNA virus) to infect plants may have been their bipartite genomic structure (Rojas et al., 2005). As with other virus families, there is a close co-evolution of Geminiviruses with their vector insects to ensure expansion and transmission, which is evidenced in the generation of symptoms that produce yellowing of the leaves of plants infected by Geminiviruses, attracting plant sucking insects and allowing the propagation of these viruses (Rojas et al., 2005).
Silencing produced by Geminiviruses: we are part of the solution
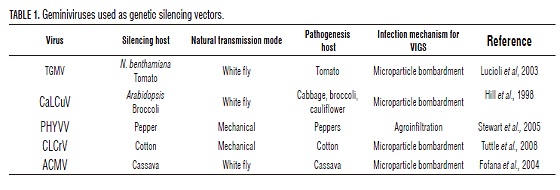
Although Geminiviruses are made up of DNA and do not present RNA intermediaries during their replication, they can induce PTGS, as initially observed when using Tomato golden mosaic virus (TGMV) as vector to silence reporter genes (Kjemtrup et al., 1998). Then came reports of virus-induced gene silencing (VIGS) in tomato (Lucioli et al., 2003), as in Geminiviruses infecting cassava like the African cassava mosaic virus Cameroon (ACMV-CM), the East African cassava mosaic Cameroon/Uganda (EACMC/ UG/V), the Sri-Lanka cassava mosaic virus (SLCMV), and the Indian cassava mosaic virus (ICMV) (Patil and Fauquet, 2009). Currently, the VIGS strategy is being used in several plants (Tab. 1).
Several hypotheses suggest how Geminiviruses could induce GS. The most accepted of these states that bidirectional transcription of transcripts produced by regions with opposing polarity would produce overlaps that would be recognized by Dicer as dsRNAs (Vanitharani et al., 2005).
Geminiviruses could induce or suppress GS. The Potato virus X (PVX) TrAP protein (Voinnet et al., 1999) was one of the first suppressors identified. Then C2, from certain Begomoviruses and AC4 from ACMV-CM, and SLCMV were characterized as PTGS suppressors (Vanitharani et al., 2004), given that transgenic plants expressing AC2 or C2 were more susceptible to the virus (Rojas et al., 2005).
Geminiviruses are used as vectors to carry out VIGS. Some examples are Tobacco curly shoot virus (TbCSV), Tomato yellow leaf curl China virus (TYLCCNV), Tomato golden mosaic virus (TGMV), and Cabbage leaf curl virus (CaLCuV) (Peele et al., 2001; Turnage et al., 2002; Tuttle et al., 2008; Golenberg et al., 2009). These vectors offer some advantages over RNA-based viruses: high sequence conservation among Geminiviruses permits adapting a virus to be used in VIGS through information from another related virus, greater genetic stability with respect to vectors based on RNA virus and can infect meristematic tissue (Padmanabhan and Dinesh-Kumar, 2008). They also have a broader range of hosts, including plants of agronomic interest (Golenberg et al., 2009). There are disadvantages for Geminiviruses as VIGS inducers: symptoms can be present in the inoculated plants, it is necessary to co-inoculate plasmids with the A and B genomes (Padmanabhan and Dinesh-Kumar, 2008) and the size of the inserts the geminiviral vectors can host is small. Nevertheless, to trigger silencing big fragments are not necessary (100 base pairs may be enough) (Vanitharani et al., 2005).
Geminiviruses and cassava
Cassava (Manihot esculenta) originated in the Amazon River basin and constitutes the nutritional base for almost one billion people (FAOSTAT, 2009). Cassava is attacked by Cassava mosaic geminiviruses (CMGs) causing the Cassava mosaic disease (CMD) (Patil and Fauquet, 2009). CMD is transmitted by the Bemisia tabaci whitefly complex (Legg and Fauquet, 2004), with isolates of such in Africa and India (Maruthi et al., 2002). The genome of the virus responsible for this disease, African cassava mosaic virus (ACMV) was sequenced in 1983 (Stanley and Gay, 1983). CMD has been reported in at least nine African countries, causing losses near US$2-billion (Legg and Fauquet, 2004). Recently, various African and Indian strains have been identified and classified (Patil and Fauquet, 2009).
CMD is found in Africa and India but not in America, although the vector, the white fly (Bemisia tabaci) is present in the Old and the New Worlds. It is possible the fly's specific biotype (B) cannot colonize cassava in America (Carabali et al., 2005). Cassava arrived to the African continent in the 16th century (Cock, 1982). It is possible that the Geminivirus, already present in this continent and infecting other types of hosts, took advantage of the extension of the cassava cultivation in Africa for its colonization. CMD expansion in Africa has been facilitated by the use of susceptible varieties, the exchange of material among geographical regions and the exchange among geminiviruses (Patil and Fauquet, 2009). One of the influential factors in the expansion of CMGs is the high diversity they present in the A genome due to the frequent recombination processes between strains and species (Fondong et al., 2000; Patil and Fauquet, 2009), among which there is also high synergy, increasing the severity of the CMD symptoms (Fondong et al., 2000).
Genetic composition
CMGs belong to the Begomovirus genus, Geminiviridae family; they have two molecules of single-strand DNA, A and B forms (Patil and Dasgupta, 2006). The A-DNA presents six open reading frames, each one for a specific protein. AC1 is the protein associated with replication (Rep), AC2 is the transcriptional activator protein (TrAP). Gene AC3 codifies for a protein that enhances replication (REn). Finally, AC4 is a silencing suppressor (Patil and Dasgupta, 2006). In the complementary sense, there are AV1 and AV2 codifying for capsid proteins. In B-DNA there is BV1 codifying nuclear-shuttle protein (NSP) and BC1 codifying movement protein (MP) implied in intraand- intercellular movement (Pita et al., 2001).
The A and B components of Begomoviruses have a common region of approximately 200 nucleotides with 80% identity (Harrison and Robinson, 1999), which contain several regulatory elements and "iteron" copies, junction sites for the protein associated to replication (Rep) (Hanley- Bowdoin et al., 2000).
CMGs have used strategies that permit them to generate a high diversity for their adaptation, among which there is a high recombination rate between A and B components (Lefeuvre et al., 2009), which may be favored by the mixed infection present in cassava cultivations where different CMG strains or species can cohabit in the same plant (Patil and Fauquet, 2009).
Silencing in cassava by Geminiviruses
Geminiviruses can induce GS or PTGS. Among those inducing PTGS, there are CMGs. In infected tobacco and cassava plants siRNAs associated to PTGS are produced (Chellappan et al., 2004). For some viruses, like ACMV and SLCMV, there is the recovery phenomenon (Rodríguez-Negrete et al., 2009), which has been correlated with PTGS and with siRNA production (Padmanabhan and Dinesh-Kumar, 2008). However, in other cases, although siRNAs have been identified, the plants do not show recovery, suggesting that there must be other viral or suppressor genetic factors that could counter silencing (Patil and Fauquet, 2009)
Most siRNAs identified correspond to the C-terminal of AC1 that overlaps with the N-terminal region of the AC2 gene from the A-DNA component. In some cases, siRNAs from the BC1 gene of the B-DNA component have been detected (Chellappan et al., 2004).
VIGS in cassava: silencing the genome!
The VIGS system has been utilized with CMGs for functional validation of genes in cassava. Fofana et al. (2004) introduced a multi-cloning region (MCS) within the ORF codifying for the capsid protein of the genome A component of ACMVCM; subsequently, it was subcloned in pBluescript to generate a silencing construct. To validate the correct functioning of silencing in cassava, phytoene desaturase (PDS) and sulfur (su) genes were used (Fofana et al., 2004) whose silencing phenotype is easily visible, plants showing bleaching and/ or yellowing, respectively (Kumagai et al., 1995). From Nicotiana benthamiana DNA constructions were made of these genes within the ACMV vector (ACMV+SU and ACMV+PDS, respectively) and cassava 60444 variety and tobacco plants were infected. In tobacco photo-bleaching was observed in plants silenced with PDS; unlike cassava did not show photo-bleaching (Fofana et al., 2004). By using the su gene, yellowing was observed in cassava and tobacco leaves, revealing that the VIGS system using ACMV works adequately (Fofana et al., 2004). To demonstrate that the system works for gene validation, Fofana et al. tested the silencing of CYP79D2, a gene involved in linamarin byosynthesis, and through RT-PCR they verified the CYP79D2 and CYP79D1 silencing, which shares 89% identity with CYP79D. Additionally, linamarin content was reduced by close to 70% in cassava leaves silenced plants (Fofana et al., 2004).
Recently, we used the VIGS strategy in cassava to silence RXam1, a candidate gene for resistance to vascular cassava bacterial blight. The ACMV-RXAM1 construct was generated and gene silencing was sought in varieties 60444 and SG107-35. Although yellowing was clearly observed in plants from the 60444 variety, only one of seven plants from the SG107-35 variety revealed a clear phenotype, suggesting that VIGS can be dependent on the variety of the plant used (Cortés and López, 2010).
Conclusions
Geminiviruses are among the main limitations in the production of several agricultural importance crops, including cassava. Plants activate GS in response to Geminiviruses, which has been used for functional validation of genes. This strategy has been implemented in cassava and has proven to be a useful tool in this plant, as well as being a great aid in the new era of functional genomics in cassava, with the recent release of its complete genome sequence (http://www. phytozome.net/cassava). ACMV has not been reported in America and for bio-security, these kinds of strategies can only be carried out in countries where CMD is present or in regions of the world where cassava is not cultivated and under strict control measures. It would be interesting to try to establish if from the viruses infecting cassava in the American continent, which present a very low incidence, viral vectors could be developed for VIGS in cassava.
Acknowledgements
We thank Doctors Claude Fauquet, Patil Basavaprabhu, and Nigel Taylor for the training in the application of the VIGS system in cassava. We also thank the anonymous evaluators for the critical revision of the manuscript. This work was sponsored by Research División Bogota at Universidad Nacional de Colombia (project No. 7915-2008): "Validación de genes candidatos de resistencia a la bacteriosis vascular en yuca mediante VIGS".
Literature cited
Bao, N., K.W. Lye, and M.K, Barton. 2004. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7, 653-662. [ Links ]
Baulcombe, D. 2005. RNA silencing. Trends Biochem. Sci. 30, 290-293. [ Links ]
Berns, K., E.M. Hijmans, J. Mullenders, T.R. Brummelkamp, A. Velds, M. Heimerikx, R.M. Kerkhoven, M. Madiredjo, W. Nijkamp, B. Weigelt, R. Agami, W. Ge, G. Cavet, P.S. Linsley, R.L. Beijersbergen, and R. Bernards. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431-437. [ Links ]
Bernstein, E., A.A. Caudy, S.M. Hammond, and G.J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363-366. [ Links ]
Bortolamiol, D., M. Pazhouhandeh, K. Marrocco, P. Genschik, and V. Ziegler-Graff. 2007. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615-1621. [ Links ]
Boutros, M., A.A. Kiger, S. Armknecht, K. Kerr, M. Hild, B. Koch, S.A. Haas, R. Paro, and N. Perrimon. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832-835. [ Links ]
Boutros, M. and J. Ahringer. 2008. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9, 554-566. [ Links ]
Briddon, R., J. Brown, E. Moriones, J. Stanley, M. Zerbini, X. Zhouy, and C.M. Fauquet. 2008. Recommendations for the classification and nomenclature of the DNA-b satellites of begomoviruses. Arch. Virol. 153, 763-781. [ Links ]
Brodersen, P. and O. Voinnet. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268-280. [ Links ]
Brodersen, P. and O. Voinnet. 2009. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10, 141-148. [ Links ]
Brown, J.K. 2008. Plant Resistance to viruses: Geminiviruses. pp. 52-58. In: Mahy, B.W.J, and M.H.V Van Regenmortel (eds.). Desk encyclopedia of plant and fungal virology. Academic Press, San Diego, CA. [ Links ]
Carabali, A., A. Bellotti, J. Montoya-Lerma, and M. Cuellar. 2005. Adaptation of Bemisia tabaci biotype B (Gennadius) to cassava, Manihot esculenta (Crantz). Crop Prot. 24, 643-649. [ Links ]
Chan, S.W., I.R. Henderson, and S.E Jacobsen. 2005. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 351-360. [ Links ]
Chapman, E.J. and J.C. Carrington. 2007. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884-896. [ Links ]
Chellappan, P., R. Vanitharani, J. Pita, and C.M. Fauquet. 2004. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 78, 7465-7477. [ Links ]
Chen, K., and N. Rajewsky. 2007. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 8, 93-103. [ Links ]
Cock, J.H. 1982. Cassava: a basic energy source in the tropics. Science 218, 755-762. [ Links ]
Cogoni, C., and G.Macino. 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa.Proc. Natl. Acad. Sci. USA 94,8854-8859. [ Links ]
Cortés, S. and C. López. 2010. Estrategia de silenciamiento génico en yuca para la validación de genes de resistencia. Acta Biol. Colomb. 15(2), 1-23. [ Links ]
Deleris, A., J. Gallego-Bartolome, J. Bao, K.D. Kasschau, and J.C. Carrington. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68-71. [ Links ]
Ding, S.W. and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130, 413-426. [ Links ]
Dorsett, Y. and T. Tuschl. 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug. Discov. 3, 318-329. [ Links ]
Eamens, A., M.B. Wang, N.A. Smith, and P.M. Waterhouse. 2008. RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol. 147, 456-468. [ Links ]
FAOSTAT. 2009. FAO database. Food and Agriculture Organization of the United Nations, http://faostat.fao.org/site/567/Desktop- Default.aspx?PageID=567#ancor; consulted: December, 2010. [ Links ]
Fargette, D., G. Konaté, C. Fauquet, E. Muller, M. Peterschmitt, and J.M. Thresh. 2006. Molecular ecology and emergence of tropical plant viruses. Annu. Rev. Phytopathol. 44, 235-60. [ Links ]
Fire, A., S. Xu, M.K. Montgomery, S.A. Kostas, and S.E. Driver. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [ Links ]
Fofana, I.B., A. Sangare, R. Collier, C. Taylor, and C.M. Fauquet. 2004. A Geminivirus-induced gene silencing system for gene function validation in cassava. Plant Mol. Biol. 56, 613-624. [ Links ]
Fondong, V.N., J.S. Pita, M.E.C. Rey, A. de Kochko, R.N. Beachy, and C.M. Fauquet. 2000. Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81, 287-297. [ Links ]
Golenberg, E., D. Sather, L. Hancock, K. Buckley, and N. Villafranco. 2009. Development of a gene silencing DNA vector derived from a broad host range Geminivirus. Plant Methods 5, 9. [ Links ]
Grimm, S. 2004. The art and design of genetic screens: mammalian culture cells. Nat. Rev. Genet. 5, 179-189. [ Links ]
Hamilton, A.J. and D.C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950-952. [ Links ]
Hammond, S.M, E. Bernstein ,D. Beach, and G.J.Hannon. 2000.An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293-296. [ Links ]
Hammond, S.M., S. Boettcher, A.A. Caudy, R. Kobayashi, and G.J. Hannon. 2001. Argonaute2, a link between genetic and Biochemical Analyses of RNAi.Science 293(5532), 1146-1150. [ Links ]
Peele, Ch., Ch.V. Jordan, N. Muangsan, M. Turnage, E. Egelkrout, P. Eagle, L. Hanley-Bowdoin, and D. Robertson. 2001. Silencing of a meristematic gene using geminivirus-derived vectors. Plant J. 27, 357-366. [ Links ]
Pita, J.S., V.N. Fondong, A. Sangare, G.W. Otim-Nape, S. Ogwal, C. M. Fauquet1. 2001. Recombination, pseudorecombination and synergism of Geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82, 655-665. [ Links ]
Pruss, G., X. Ge, X.M. Shi, J.C. Carrington, and V. Bowman. 1997. Plant viral synergism: the potyviral genome encodes a broadrange pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9, 859-868. [ Links ]
Purkayastha, A., and I. Dasgupta. 2009. Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol. Biochem. 47, 967-976. [ Links ]
Rines, H.W. 1985. Sodium azide mutagenesis in diploid and hexaploid oats and comparison with ethyl methanesulfonate treatments. Environ. Exp Bot. 25, 7-16. [ Links ]
Rodríguez-Negrete, E.A., J. Carrillo-Tripp, and R.F. Rivera- Bustamante. 2009. RNA Silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 83, 1332-1340. [ Links ]
Robertson, D. 2004. VIGS vectors for gene silencing: many targets, many tools. Annu. Rev. Plant Biol. 55, 495-519. [ Links ]
Rojas, M.R., C. Hagen, W.J. Lucas, and R.L. Gilbertson. 2005. Exploiting chinks in the plant's armor: evolution and emergence of Geminiviruses. Annu. Rev. Phytopathol. 43, 361-394. [ Links ]
Shuman, H.A., and T.J Silhavy. 2003. The art and design of genetic screens: Escherichia coli. Nat. Rev. Genet. 4, 419-431. [ Links ]
Schoelz, J.E. 2008. Caulimoviruses: general features. pp. 147-155. En: Mahy, B.W.J., and M.H.V. Van Regenmortel (eds.). Desk encyclopedia of plant and fungal virology. Academic Press, San Diego, CA. [ Links ]
Soosaar, J.L., T.M. Burch-Smith, and S.P. Dinesh-Kumar. 2005. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 3, 789-798. [ Links ]
Stanley, J., and M. Gay. 1983. Nucleotide sequence of cassava latent virus DNA. Nature 301, 260-262. [ Links ]
Stewart, C.S., B.C. Kang, K. Liu, M. Mazourek, S.L. Moore, I. Paran, M. Jahn. 2005. The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J. 42, 675-688. [ Links ]
Turnage, M.A., N. Muangsan, Ch. Peele, and D. Robertson. 2002. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30, 107-114. [ Links ]
Tuttle, J.R., A.M. Idris, J.K. Brown, C.H. Haigler, and D. Robertson. 2008. Geminivirus-mediated gene silencing from cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 148, 41-50. [ Links ]
Umbach, J.L., and B.R. Cullen. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Gene Dev. 23, 1151-1164. [ Links ]
Vanitharani, R., P. Chellappan, J.S. Pita, and C.M. Fauquet. 2004. Differential roles of AC2 and AC4 of cassava Geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78, 9487-9498. [ Links ]
Vanitharani, R., P. Chellappan, and C.M. Fauquet. 2005. Geminiviruses and RNA silencing. Trends Plant Sci. 10, 144-151. [ Links ]
Vizir, I.Y., M.L. Anderson, Z.A, Wilson, and B.J. Mulligan. 1994. Isolation of deficiencies in the Arabidopsis genome by gammairradiation of pollen. Genetics 137, 1111-1119. [ Links ]
Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206-220. [ Links ]
Voinnet, O. 2008. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13, 317-328. [ Links ]
Voinnet, O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669-687. [ Links ]
Voinnet, O., Y.M. Pinto, and D.C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA. 96, 14147-14152. [ Links ]
Waterhouse, P.M., and A.F. Fusaro. 2006. Plant science. Viruses face a double defense by plant small RNAs. Science 313, 54-55. [ Links ]
Wesley, S.V., C.A. Helliwell, N.A. Smith, M.B. Wang, D.T. Rouse, Q. Liu, P.S Gooding, S,P Singh, D. Abbott, P.A Stoutjesdijk, S.P Robinson, A.P Gleave, A G Green, P.M Waterhouse. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581-590. [ Links ]