Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Agronomía Colombiana
Print version ISSN 0120-9965
Agron. colomb. vol.32 no.2 Bogotá May/Aug. 2014
https://doi.org/10.15446/agron.colomb.v32n2.43968
http://dx.doi.org/10.15446/agron.colomb.v32n2.43968
CROP PROTECTION
1 Plant Virus Laboratory, Biotechnology Institute, Universidad Nacional de Colombia. Bogota (Colombia). mmguzmanb@unal.edu.co
2 International Center for Tropical Agriculture (CIAT ). Palmira (Colombia).
Received for publication: 11 May, 2014. Accepted for publication: 30 July, 2014.
ABSTRACT
The Potato yellow vein virus (PYVV), a Crinivirus with an RNA tripartite genome, is the causal agent of the potato yellow vein disease, reported in Colombian since 1950, with yield reductions of up to 50%. Co-infection of two or more viruses is common in nature and can be associated with differences in virus accumulation and symptom expression. No evidence of mixed infection between PYVV and other viruses has been reported. In this study, eight plants showing yellowing PYVV symptoms: four Solanum tuberosum Group Phureja (P) and four Group Andigena (A), were collected in Cundinamarca, Colombia to detect mixed infection in the isolates using next generation sequencing (NGS). The Potato virus Y (PVY) complete genome (similar to N strain) and the Potato virus V (PVV) partial genomes were detected using NGS and re-confirmed by RT-PCR. Preliminary field screening in a large sample showed that PYVV and PVY co-infect potato plants with a prevalence of 21% within the P group and 23% within the A group. This is the first report of co-infection of PYVV and potyvirus in Colombia and with the use of NGS. Considering that potyviruses enhance symptom severity and/or yield reductions in mixed infections, our results may be relevant for disease diagnosis, breeding programs and tuber certification.
Key words: viral diseases, Potato virus V, prevalence, NGS , potato, root vegetables.
RESUMEN
El Potato yellow vein virus (PYVV) es un Crinivirus de genoma RNA tripartita, agente causal de la enfermedad del amarillamiento de las nervaduras de la papa reportada desde 1950 en Colombia con reducción de producción hasta 50%. La coinfección entre virus en un mismo hospedero es común en la naturaleza y se asocia con cambios en acumulación viral y en la expresión de síntomas. Hasta el momento no se ha reportado coinfección entre PYVV y otros virus en papa. En este estudio se colectaron en Cundinamarca-Colombia ocho plantas con síntomas de PYVV: cuatro Solanum tuberosum Grupo Phureja (P) y cuatro del Grupo Andigena (A). Utilizando secuenciamiento de alto rendimiento (NGS) en los aislamientos se detectó infección mixta de PYVV con potyvirus que se reconfirmó con RT-PCR. Estudios preliminares en una muestra mayor de campo confirmaron que PYVV y PVY coinfectan papa con 21% de prevalencia para el grupo P y 23% para el grupo A. Este es el primer reporte de coinfección de PYVV y potyvirus en Colombia. Teniendo en cuenta que los potyvirus pueden incrementar la severidad de síntomas y/o reducir el rendimiento en infecciones mixtas, los resultados presentados pueden ser relevantes para el diagnóstico de enfermedades virales, programas de propagación y de certificación de semillas.
Palabras clave: enfermedades virales, Potato virus V, prevalencia, NGS , papa, hortalizas de raíz.
Introduction
Co-infection is a natural event involving different viruses or strains from the same virus being present in a host at the same time (Bennett, 1953). Such viruses interact in some cases, thereby increasing (synergism) or decreasing (antagonism) the accumulation and expression of the symptoms of one or more viruses, thus affecting crop yield (Goodman and Ross, 1974; Vance, 1991; Pruss et al., 1997). The world's potato production is affected by at least 40 different viruses (Vreugdenhil, 2007); potato-infecting viruses are spreading fast worldwide; mostly because of infected tubers being used as seed-tubers for propagating crops, thereby leading to the possibility of interaction (Davey, 2013); and also by dispersion caused by natural vectors (Salazar et al., 2000).
Mixed Potato virus X (PVX) and Potato virus Y (PVY) infection in Solanaceae sp. has been one of the most studied viral synergisms; it causes increased PVX accumulation without interfering with PVY accumulation, but can reduce yield by around 80% (Vance, 1991; Anjos et al., 1992; Vance et al., 1995). PVY (family Potyviridae, genus Potyvirus) is one of the most important potato pathogens worldwide. Its genome consists of a single-stranded, positive-sense RNA molecule of about 10 kb in length (Dougherty and Carrington, 1998; Hall et al., 1998). Gene interactions involving different strains have been characterized based on host response and resistance, including the ordinary PVY (PVYO), Tobacco venial necrosis (PVY-N) and stipple streak (PVY-C) strain groups of PVY (Schubert et al., 2007; Singh et al., 2008; Moury, 2010). The North American (NA ) isolate of the potato tuber necrotic strain of PVY (PVY-NTN ) is serologically related to PVY-N; however, PVY-NTN has been found to be a recombinant of PVY-O and PVY-N in the CP gene at the molecular level (Boonham et al., 2002). Phylogenetic analysis of CP sequences from Colombian and Chilean isolates, which had previously been characterized as PVY-NTN , has clustered them together in a new group which is closely related to PVY-N and PVY-NTN (Moury, 2010; Gil et al., 2011).
Reports of co-infection involving potyviruses and different viral groups are common. Co-infection with Comovirus in the soybean (Anjos et al., 1992), with Crinivirus in the sweet potato (Untiveros et al., 2007) and with Potexvirus in potato plants (Vance, 1991) has been reported. Previous studies have indicated that this is most likely due to P1/HCPro post-transcriptional gene silencing (PTGS ) suppressor expression which allows non-potyvirus accumulation levels to increase without being repressed by plant PTGS defences (Anjos et al., 1992; Vance et al., 1995; Pruss et al., 1997; Anandalakshmi et al., 1998).
Crinivirus co-infection with other viral groups has been reported, e.g. Potyvirus, Carlavirus, Cucumovirus, Ipomovirus and Cavemovirus (Untiveros et al., 2007; Cuellar et al., 2011). Regarding Crinivirus, Sweet potato chlorotic stunt virus (SPCSV) co-infection with the Sweet potato feathery mottle virus (SPFMV) results in increased symptom severity and yield loss in the sweet potato, which is believed to be mediated by SPCSV endoribonuclease III (RNA se3) acting as a silencing suppressor and, thus, allowing synergism with SPFMV (Untiveros et al., 2007; Kreuze et al., 2008; Cuellar et al., 2009).
PYVV is a re-emergent and quarentenary Crinivirus known as the causal agent of the Potato yellow vein disease (PYVD), affecting production in Colombia by 25 to 50% (Salazar et al., 2000; Guzmán-Barney et al., 2012), and has a prevalence of 5.6 to 11.0% in potato crops of Group P reported in three Colombian states (Franco-Lara et al., 2012). The viral genome consists of three single-stranded, positive-sense RNA s and at least two defective RNA s (Livieratos et al., 2004; Eliasco et al., 2006). Major coat protein (CP) gene studies have indicated low variability (Offei et al., 2004; Guzmán et al., 2006; Rodríguez-Burgos et al., 2009; Chaves-Bedoya et al., 2013) compared to high variability regarding the minor coat protein (mCP) gene and the homologue heat shock protein (HSP70h) gene (Chavez-Bedoya et al., 2014).
Considering the importance of PYVV in Colombia and other Andean countries and the widespread occurrence of potyviruses, such as PVY, this study aimed to detect PYVV co-infection of field potato plants with other viruses which might partly explain the leaf symptom expression variability and yield loss which have been observed by our research group in previous studies. Next-generation sequencing (NGS ) and RT-PCR were used to detect the viruses and establish co-infection. Phylogenetic analysis of the sequenced amplicons was used to identify and cluster the detected viruses. An NGS approach has thus been used for the first time to demonstrate that PYVV can co-infect potato plants along with PYV and PVV.
Materials and methods
Plant material
Eight Solanum tuberosum Group Phureja (P) (4) and Group Andigena (A) (4) plants expressing leaf yellowing symptoms were collected near Chipaque, Cundinamarca (Colombia), for RT-PCR diagnosis and NGS analysis. Leaf samples were collected at different times of the year from 61 S. tuberosum Group P (51 symptomatic and 10 symptomless) and 39 of Group A (28 symptomatic and 11 symptomless) potato crops grown in Cundinamarca, using random sampling to establish virus prevalence.
RNA and siRNA extraction
For siRNA extraction, an initial total RNA extraction was performed from 4 g symptomatic leaves from each of the eight plants using Trizol reagent (Invitrogen™, Thermo Fisher Scientific, Waltham, MA); 50 ug were separated on 4% agarose gel with microRNA markers (New England Biolabs®, Ipswich, MA) and visualised using SY BRSafe (Invitrogen™, Thermo Fisher Scientific, Waltham, MA). Bands located between 21 and 30 bp were excised and purified using a gel extraction spin column kit (Bio-Rad Laboratories, Hercules, CA). The pellet was washed with 75% ethanol and dried at room temperature, according to the procedure described by Kreuze et al. (2009). All small interference RNA s (siRNA ) were sent to Fasteris Life Science (Fasteris, Plan-les-Ouates, Switzerland) for processing and sequencing on an Illumina Genome Analyzer II (Illumina®, San Diego, CA). Total RNA for further analysis was obtained from 1 g leaves using Trizol reagent (Invitrogen™, Thermo Fisher Scientific, Waltham, MA). The RNA was purified using chloroform, precipitated with isopropanol and washed with 70% ethanol.
NGS analysis
Reads obtained by Illumina sequencing were assembled using Velvet (Zerbino, 2008) and Assembly-Assembler script software. The PVY genome sequence (NC_001616, AY 884984) was initially used as a template for aligning siRNA reads using MA Q software (Redmond, WA). Different contigs were produced depending on the software used and different parameters, as published elsewhere (Flores et al., 2011). Larger contigs were assembled using SeqMan (DNASTA R software, Madison, WI), combining the consensus contigs and sequences produced using MA Q and Velvet. The assembled contigs were compared with the NCBI nucleotide and protein databases using the NCBI BLAST ® (Bethesdam, MD) database search tool (Altschul et al., 1997); their translated peptides were matched to the corresponding viruses in each plant. Virus-specific contig coverage and distribution by siRNA was determined using MA Q (default parameters) and the results were exported to R statistical software (version 2.13.0) and Microsoft Excel® for further analysis.
PYVV and PYV coat protein gene amplification
Two hundred ng total RNA were denatured at 72°C for 10 min and chilled on ice for 2 min to confirm the presence of PYVV and the potyviruses. RNA was reverse-transcribed for 1 h at 42°C in the presence of 0.4 mM of the corresponding reverse primer (Tab. 1), 1X reaction buffer (Epicentre, Illumina®, San Diego, CA), 1 mM dNT Ps (Bioline, London), 10 mM DTT (Epicentre, Illumina®, San Diego, CA), 1.6U RNA se inhibitor (Fermentas, Thermo Fisher Scientific, Waltham, MA ), and 8U MM LV HP (Epicentre, Illumina®, San Diego, CA) to 10 mL final volume. Final denaturing took place at 72°C for 10 min.
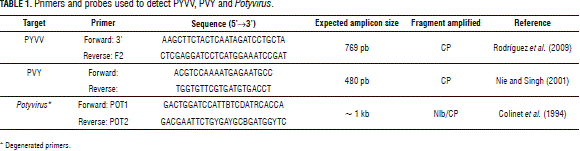
PCR was carried out using 1.6 mL of the RT-PCR product diluted five times in 1X buffer NH4 (Bioline, London), 2.0 mM MgCl2 (Bioline, London), 0.4 mM dNT Ps (Bioline, London), 0.4 mM of each primer (Tab. 1) and 1U of Biolase (Bioline, London) to 10 mL final volume. Samples were initially denatured at 94°C for 4 min and 35 cycles of PCR with 30 s denaturing at 94°C, 30 s of annealing at 60°C and 30 s of extension at 72°C. The series of cycles was followed by a final extension step for 10 min at 72°C.
Seven mL of each PCR product were analyzed on a 2% agarose gel in TA E buffer, stained with SY BRSafe (Invitrogen™, Thermo Fisher Scientific, Waltham, MA ), ran for 45 min at 70 V and photographed with a gel digitalizer (BioRad). Amplicons obtained with the potyvirus degenerate primers were cloned in PCR 4-TOPO (Invitrogen™, Thermo Fisher Scientific, Waltham, MA ) and recombinant plasmids were purified from E. coli using a Quicklyse miniprep kit (Quiagen, Hilden, Germany). Plasmid preparations were sequenced using both forward and reverse primers. Macrogen, Korea, did the sequencing.
Sequence alignment and phylogenies
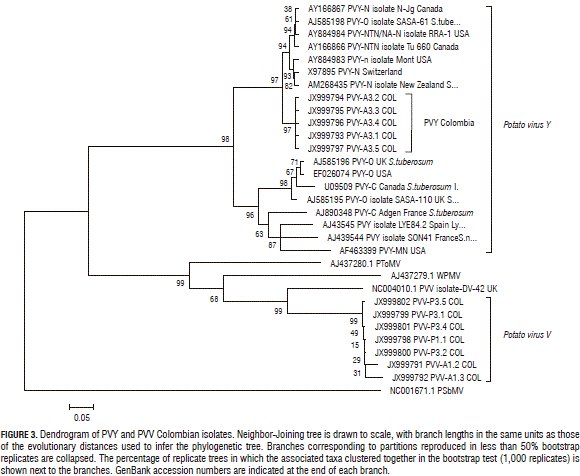
To confirm the presence of PVY and PVV in the NGS analyzed samples, isolates of PYV and PVV were amplified with the degenerated primers POT1 and POT2 (Tab. 1). Amplicons were cloned and sent to Macrogen (Seoul, Korea) for Sanger sequencing. Molecular Evolutionary Genetics Analysis (MEGA) software (version 4.0) (Tamura et al., 2007) was used for bioinformatics analysis and Clustal W for aligning sequences (Thompson et al., 1994). The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al., 2004), units being the number of base substitutions per site. Evolutionary history was inferred using the Neighbour-Joining method (Saitou and Nei, 1987). Statistical confidence was evaluated using a bootstrap test with 1,000 replicates (Felsenstein, 1985). Pea seed-borne mosaic virus (PSbMV) was used as the outgroup.
Results
NGS analysis
Eight leaf samples of PYVD symptomatic Solanum tuberosum Groups P and A plants were collected from crops in Cundinamarca, Colombia. siRNA s were purified from total RNA extracts and sent for NGS using the Illumina platform. Reads between 21 to 24 nt were used to assemble contigs; 0.2 to 1.2 million reads were obtained, having a higher proportion of host and unspecific siRNA s. Potyviral sequences accounted for 63% of the reads for virus-specific siRNA s, particularly represented by 21 and 22 nt siRNA s. While PYVV sequences accounted for 8% of the 21 and 22 nt siRNA s, the remaining contigs corresponded to sequences with similarity to sequences for other viral families, host sequences or unknown sequences.
The reads were assembled into contigs; Tab. 2 shows those having over 70% similarity with other viruses. Several contigs were found having similarity with virus sequences from the families Betaflexiviridae, Caulimoviridae, Closteroviridae and Potyviridae. However, most contigs were discarded due to the contig length (less than 100 nt) or the contig amount (one to three contigs) (Tab. 2), leading to not taking into account the family Caulimoviridae and some members of the family Potyviridae as candidate viruses.
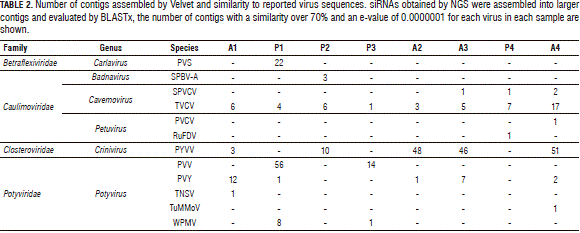
Regarding Cavemovirus, Turnip vein clearing virus (TVCV) was also discarded as a candidate due to Cavemovirus genome similarity to sequences integrated in the host genome (Lockhart et al., 2000) and all the contigs found only matched open reading frames (ORF) 3 and 4.
The remaining candidate viruses were Potyvirus PVV and PVY. Most of the larger contigs had high similarity with sequences reported for PVY which lead to the complete assembly of sequences corresponding to the virus genome (Fig. 1A).
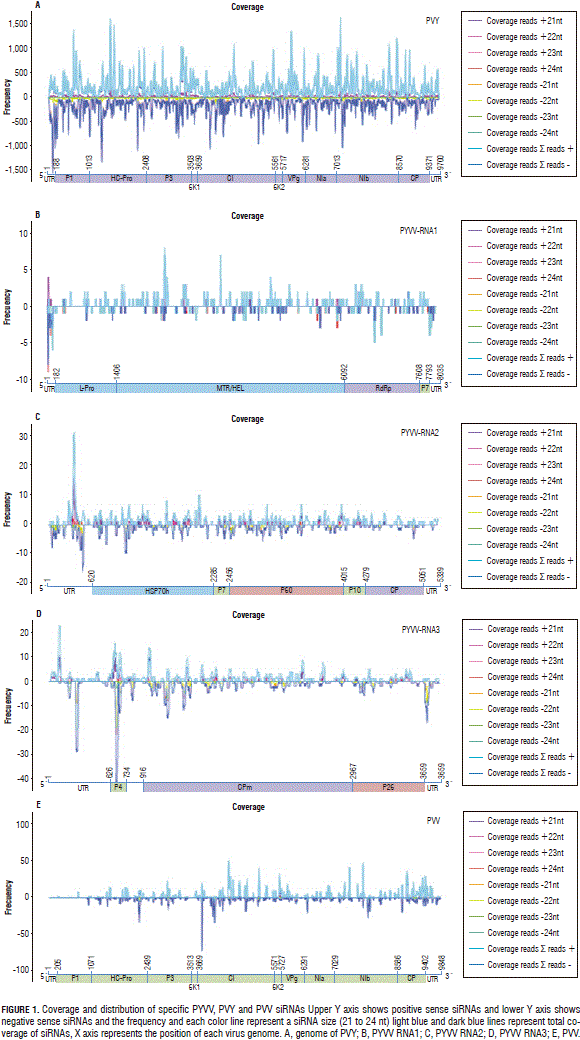
MAQ software was used for determining read frequency and distribution in the genome for both potyviruses and PYVV. Even when several contigs having high identity were obtained for PYVV and PVV, siRNA frequency was less than 50 reads for each position in most of the genome, so complete sequences could not be assembled (Fig. 1B to 1D). The results gave less than 50 reads for each point in the PVV and PYVV genome; whereas, frequency at each point was around 1,500 reads for PVY (Fig. 1).
The reads led to assembling 46% of the first PYVV RNA and 76% of the second and third RNA s; potyvirus coverage was 99% for PVY and 50 for PVV; coverage and frequency were lower at the 5' end than the 3' end for PVV.
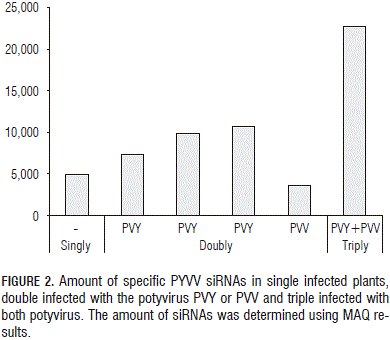
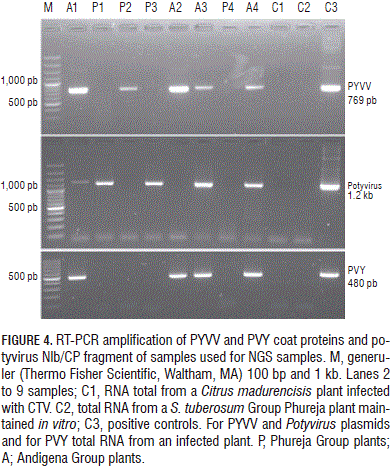
The amount of each virus-specific siRNA (Fig. 1) showed that PVY siRNA s were 17 times more abundant than those for PYVV in dual-infected plants; PVV-specific reads were four times less abundant than PYVV-specific siRNA s in dual-infected plants while PYVV siRNA s were 1.6 times more abundant in single-infected plants than those for PVV. The amount of PYVV-specific siRNA s was almost twice as high when co-infecting with PVY and four times higher when co-infecting with PVY and PVV simultaneously (Fig. 2). This contrasted with the RT-PCR and sequencing results which confirmed the presence of PVV and PYVV (Fig. 3) even though the number of reads for these viruses was under-represented in the NGS data set as compared to PVY (Tab. 2, Fig. 4).
Sequence analysis of samples A1, P1 and P3 (Fig. 4) revealed similarity to PVV, thereby corroborating the results shown in Tab. 2. It also showed that the Potyvirus degenerate primers allowed PVY to be detected in just one of the tested samples.
PYVV and PVY Prevalence evaluation
The aforementioned results showed PYVV and PVY simultaneously infecting the same plant sample. A preliminary prevalence assay was made to corroborate co-infection in the field. Symptomatic and symptomless Group P (61 plants) and A (9 plants) samples were collected in Chipaque, Cundinamarca, and tested with specific primers. RT-PCR was used for estimating the number of singly- and duallyinfected plants (Fig. 5). Results revealed that 21% of Group P and 23% of Group A samples were dual-infected and that just 15% of A samples and 16% of P samples were not infected by any of the viruses being tested.
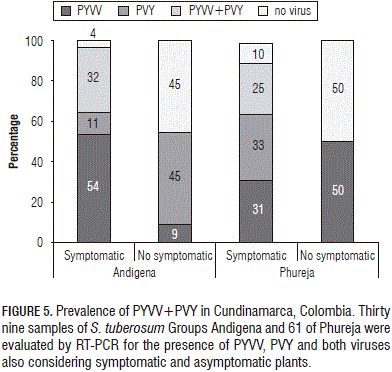
Eleven symptomless plants were collected in the Group A and the percentages of singly- and dually-infected plants were calculated. The results showed that most PYVV singlyinfected plants were symptomatic (54%) whereas most PVY singly-infected plants were symptomless (45%); all duallyinfected plants evaluated here exhibited symptoms (Fig. 5).
Ten symptomless plants were collected in the Group P. The results showed that all symptomless plants were either PYVV singly-infected (50%) or were not infected by any of the viruses being evaluated (50%); whereas 25% of the symptomatic plants were dually-infected and similar amounts of singly-infected PYVV and PVY were found (Fig. 6).
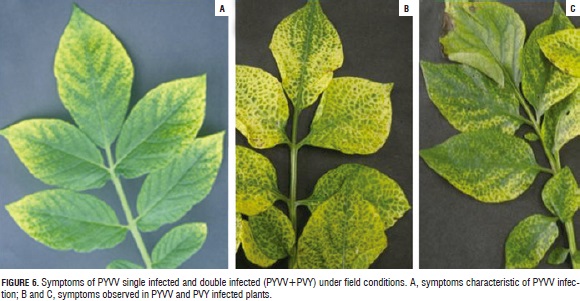
All dually-infected plants evaluated here exhibited symptoms (Fig. 5) and their expression was documented; singly-infected plants exhibited typical symptoms of PYVV infection, meaning yellow leaves having green veins (Salazar et al., 2000). Dually-infected plants showed mild and severe mosaic with inter-venial yellowing in some cases; this differed greatly from previously-described symptoms caused by PYVV (Fig. 6).
Discussion
PYVV is a re-emergent Crinivirus becoming one of the most important potato viruses in Andean countries where the infection is responsible for 25 to 50% yield reduction (Salazar et al., 2000; Franco-Lara et al., 2009; Guzmán- Barney et al., 2012).
Host co-infection by different viruses may cause increase in symptom severity and yield loss (Goodman and Ross, 1974; Vance, 1991; Pruss et al., 1997). Dual PVY and PVX infection is responsible for large potato losses around the world, accounting for a 3- to 10-fold increase in symptom severity according to tobacco plant observations (Vance, 1991); nevertheless, co-infection does not always result in increased symptoms and/or yield loss. Some viruses interact without affecting each another (i.e. neutralism) (Bennett, 1953).
It is well known that virus interactions could affect virus accumulation, symptom expression and crop yield; for that reason, in the present study, next generation sequencing approach (NGS) was used to detect different viruses that could co-exist with PYVV in field potato plants and that may be responsible for some of the symptom expression differences observed in field-collected plants (Franco-Lara et al., 2009).
Between 0.2 and 1.2 million 21 to 24 nt RNA reads were obtained and used to construct larger contigs which were compared to other GenBank sequences using BLAST X. Several contigs having identity with several plant virus families were found; however, closer analysis using different parameters (similarity, e-value, contigs length, etc.) suggested that a large number of contigs had been misidentified (e.g. viruses in the families Luteroviridae, Begomoviridae, Masteroviridae, Tymoviridae and Virgaviridae).
The larger contigs matched PVY sequences, and the length and amount led to the assembly of full-length sequences that were used to perform phylogenetic analysis (Fig. 3). Those showed that the Colombian samples' PVY sequences grouped together with European and North-American NTN strain isolates, although they were not identical to those from previous reports (Gil et al., 2011). Interestingly, Potato virus S (PVS) was detected in a P group sample. PVS has already been reported in Colombia in single (Sánchez de Luque et al., 1991; Gil et al., 2011) and mixed infections with PLRV, PVX, PVS and PVY (Guzmán et al., 2010).
Preliminary studies for PYVV and PVV co-infection have been reported previously (Rodríguez et al., 2011). Full-length genome sequences could not be formed for PYVV and PVV (<76% and 51% coverage, respectively); Fig. 1 shows that the number of PYVV siRNA s obtained was 17-fold lower than that obtained for PVY and only 1.6 higher than that obtained for PVV. PYVV-specific siRNA s were twice as high when co-infecting with PVY and four times higher when co-infecting with both PVY and PVV (Fig. 2). Larger contigs were obtained for PVY and up to 99% of the genome was assembled.
Contigs similar to Cavemovirus TVCV were found in all the analyzed samples (Tab. 2); nevertheless, this could have been due to the presence of pararetrovirus sequences integrated in the host genome, which has been already been described in other Solanaceae (Lockhart et al., 2000).
A triple co-infection was noticed in one of the Group P samples analyzed here (this study's scope precluded further investigation). Triple infection including Crinivirus has already been reported as causing greater symptom severity than that in dual-infection (Untiveros et al., 2007); the effects and prevalence of PVY, PVV and PYVV triple infection should thus be assessed.
A group of 100 field potato plants was used for preliminary estimation of dual-infection (PYVV-PVY) prevalence; up to 23% of the samples analysed here were dually-infected. In a screening done in Group P crops in three Colombian states during 2008, PYVV was reported as being present in 5.6 to 11% of the symptomatic samples and in 25% of the symptomless plants (Franco-Lara et al., 2012); PVY prevalence was estimated at around 72% (Gil et al., 2011). Prevalence analysis revealed that all dually-infected plants exhibited symptoms; in contrast, singly-infected plants in the Group A only had symptoms in 54% of the cases for PYVV and 45% for PVY and the Group P had 56% PYVV and 33% PVY (Fig. 5).
PYVV singly-infected plants showed typical symptoms: leaflets having clearing of the secondary and tertiary veins and also intense yellow leaflets having green central veins (Salazar et al., 2000); whilst symptoms for the duallyinfected plants (PVY-PYVV) included mild and severe mosaic, with yellow central veins in some cases (Fig. 6). Changes in symptom expression have been observed in virus interactions, namely Lettuce infectious yellows virus (LIYV, Crinivirus) and Turnip mosaic virus (TuMV, Potyvirus) or Tomato chlorosis virus (ToCV, Crinivirus) (Wang et al., 2009) and Tomato spotted wilt virus (TSWV, Tospovirus, Bunyaviridae) (García-Cano et al., 2006), whose interactions cause increased symptom severity leading to host death in some cases.
Potyvirus co-infection with non-related viruses can result in synergism, usually over-accumulation of non-Potyvirus, without affecting Potyvirus levels (Vance et al., 1995). It was established that PYVV co-infects with PVY and also possibly with PVV. Helper component proteinase (HCPro) is known to act as a silencing suppressor in Potyvirus, thereby affecting siRNA accumulation (Mallory et al., 2001) and explaining how PVY could allow other viruses to increase their effect in a particular host. By promoting higher virus accumulation, synergism could indirectly affect symptom severity and the efficiency of transmission by insect vectors. Types of Crinivirus have also been found to have a synergistic effect on unrelated viruses; when SPSCV (Crinivirus) interacts with SPFMV (Potyvirus) accumulation levels have not differed from single infected plants but SPFMV has increased its levels up to 600-fold and this has correlated with intensified symptom severity (Karyeija et al., 2000). Such synergism is believed to be mediated by the SPCSV RNase3 protein which acts as a silencing suppressor (Kreuze et al., 2008; Cuellar et al., 2009). It would be interesting to identify the silencing suppressor protein in PYVV and ascertain its role in PYVV co-infection.
Preliminary assays were conducted using qPCR and ELISA tests; however, no PYVV and PVY synergistic interaction was detected; results that may be due to the reduced number of samples used and the lack of a follow-up study, which led us to suggest that PYVV may have been interacting with PVY. Further studies are required to ascertain this. The results are very important and should be taken into account by phyto-sanitary institutions. The prevalence study and symptom detection represent preliminary steps and should be enforced by research on a larger scale.
Ackwonlegments
We thank J. Kreuze and M. Florez from the International Potato Center for expert advice in small RNA analysis and A. Hernandez from the Biotechnology Institute of the National University of Colombia for RT-qPCR advice. This work was supported by TWAS -UNESCO-ICGEB and Colciencias project N. 202010016358.
Literature cited
Altschul, S.F., T.L. Madden, A.A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D.J. Lipman. 1997. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [ Links ]
Anandalakshmi, R., G.J. Pruss, X. Ge, R. Marathe, A.C. Mallory, T.H. Smith, and V.B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079-13084. [ Links ]
Anjos, J.R., U. Järlfors, and S.A. Ghabrial. 1992. Soybean mosaic potyvirus enhances the titer of two cucumoviruses in dually infected soybean plants. Phytopathol. 82, 1022-1027. [ Links ]
Bennett, C. 1953. Interactions between viruses and virus strains. Adv. Virus Res. 1, 40-67. [ Links ]
Boonham, N., K. Walsh, M. Hims, S. Preston, J. North, and I. Barker. 2002. Biological and sequence comparisons of Potato virus Y isolates associated with potato tuber necrotic ringspot disease. Plant Pathol. 51, 117-126. [ Links ]
Chaves-Bedoya, G., M. Guzmán-Barney, and L. Ortíz-Rojas. 2013. Genetic heterogeneity and evidence of putative darwinian diversifying selection in Potato yellow vein virus (PYVV). Agron. Colomb. 31, 161-168. [ Links ]
Chaves-Bedoya, G., K. Cubillos, and M. Guzmán-Barney. 2014. First report of recombination in Potato yellow vein virus (PYVV) in Colombia. Trop. Plant Pathol. 39, 234-241. [ Links ]
Colinet, D., P. Kummert, P. Lepoivre, and J. Semal. 1994. Identification of distinct Potyviruses in mixedly-infected sweetpotato by the polymerase chain reaction with degenerate primers. Phytopathology 84, 65 69. [ Links ]
Cuellar, W.J., J.F. Kreuze, M.L. Rajamäki, K.R. Cruzado, M. Untiveros, and J.P. Valkonen. 2009. Elimination of antiviral defense by viral RNase III. Proc. Natl. Acad. Sci. USA 106, 10354-10358. [ Links ]
Cuellar, W.J., J. De Souza, I. Barrantes, S. Fuentes, and J.F. Kreuze. 2011. Distinct cavemoviruses interact synergistically with Sweet potato chlorotic stunt virus (genus Crinivirus) in cultivated sweet potato. J. Gen. Virol. 92, 1233-1243. [ Links ]
Davey, T., S.F. Carnegie, G.S. Saddler, and W.J. Mitchell. 2013. The importance of the infected seed tuber and soil inoculum in transmitting Potato mop-top virus to potato plants. Plant Pathol. 63, 88-97. [ Links ]
Dougherty, W.G. and J.C. Carrington. 1998. Expression and function of potyviral gene products. Annu. Rev. Phytopathol. 26, 123-143. [ Links ]
Eliasco, E., I.C. Livieratos, G. Müller, M. Guzmán, L.F. Salazar, and R.H.A. Coutts. 2006. Sequence of defective RNA s associated with potato yellow vein virus. Arch. Virol. 151, 201-204. [ Links ]
Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783-791. [ Links ]
Flores, M., R. Silvestre, J. Kreuze, and W. Cuellar. 2011. Evaluación de material indexado para virus por secuenciamiento a gran escala. Fitopatol. Colomb. 35(suppl.), 60. Memorias XXX Congreso Colombiano y XV Latinoamericano de Fitopatología. Bogota. [ Links ]
Franco-Lara, L., A. Villamil, A. Guateque, and M. Guzman. 2009. Evidence of latency of PYVV in tubers and plants of Solanum phureja. Phytopathol. 99, S37. [ Links ]
Franco-Lara, L., D. Rodríguez, and M. Guzman-Barney. 2012. Prevalence of Potato yellow vein virus (PYVV) in Solanum tuberusum Group Phureja fields in three states of Colombia. Amer. J. Pot. Res. 90, 324-330. [ Links ]
García-Cano, E., R.O. Resende, R. Fernández-Muñoz, and E. Moriones. 2006. Synergistic interaction between Tomato chlorosis virus and Tomato spotted wilt virus. Phytopathol. 96, 1263-1269. [ Links ]
Gil, J.F., J.M. Cotes, and M. Marin. 2011. Incidencia de potyvirus y caracterización molecular de PVY en regiones productoras de papa (Solanum tuberosum L.) de Colombia. Rev. Colomb. Biotecnol. 13, 85-93. [ Links ]
Goodman, R.M. and A.F. Ross. 1974. Enhancement of Potato Virus X synthesis in doubly infected tobacco occurs in doubly infected cells. Virology 58, 16-24. [ Links ]
Guzmán, M., E. Ruiz, N. Arciniegas, and R.H.A. Coutts. 2006. Occurrence and variability of Potato yellow vein virus in three departments of Colombia. J. Phytopathol. 154, 748-750. [ Links ]
Guzmán, M., V. Román, L. Franco, and P. Rodríguez. 2010. Presencia de cuatro virus en algunas accesiones de la Colección Central Colombiana de papa mantenida en campo. Agron. Colomb. 26, 225-234. [ Links ]
Guzmán-Barney, M., L. Franco, D. Rodríguez, L. Vargas, and J. Fierro. 2012. Yield losses in Solanum tuberosum Group Phureja cultivar Criolla Colombia in plants with symptoms of PYVV in field trials. Amer. J. Pot. Res. 89, 438-447. [ Links ]
Hall, J.S., B. Adams, T.J. Parsons, R. French, L.C. Lane, and S.G. Jenson. 1998. Molecular cloning, sequencing and phylogenetic relationship of a new potyvirus: sugarcane streak mosaic virus, and a re-evaluation of the classification of the potyviridae. Mol. Phylogenet. Evol. 10, 323-332. [ Links ]
Karyeija, R.F., J.F. Kreuze, R.W. Gibson, and J.P. Valkonen. 2000. Synergistic interactions of a potyvirus and a phloem-limited crinivirus in sweet potato plants. Virology 269, 26-36. [ Links ]
Kreuze, J.F., I. Samolski Klein, M. Untiveros Lazaro, W. Cuellar Chuquiyuri, G. Lajo Morgan, P.G. Cipriani Mejía, M. Ghislain, and J.P.T. Valkonen. 2008. RNA silencing-mediated resistance to a crinivirus (Closteroviridae) in cultivated sweet potato (Ipomoea batatas L.) and development of sweet potato virus disease following co-infection with a potyvirus. Mol. Plant Pathol. 9, 589-598. [ Links ]
Kreuze, J., A. Perez, M. Untiveros, D. Quispe, S. Fuentes, I. Barker, and R. Simon. 2009. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNA s: A generic method for diagnosis, discovery and sequencing of viruses. Virology 388, 1-7. [ Links ]
Livieratos, I.C., E. Eliasco, G. Müller, R.C. Olsthoorn, L.F. Salazar, C.W. Pleij, and R.H. Coutts. 2004. Analysis of the RNA of Potato yellow vein virus: evidence for a tripartite genome and a conserved 3'-terminal structures among members of the genus Crinivirus. J. Gen. Virol. 85, 2065-2075. [ Links ]
Lockhart, B.E., J. Menke, G. Dahal, and N.E. Olszewski. 2000. Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J. Gen. Virol. 81, 1579-1585. [ Links ]
Mallory, A.C., L. Ely, T.H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V.B. Vance. 2001. HC-Pro suppression of transgene silencing eliminates the small RNA s but not transgene methylation or the mobile signal. Plant Cell 13, 571-584. [ Links ]
Moury, B. 2010. A new lineage sheds light on the evolutionary history of Potato virus Y. Mol. Plant Pathol. 11, 161-168. [ Links ]
Nie, X. and R.P. Singh. 2001. A novel usage of random primers for multiplex RT-PCR detection of virus and viroid in aphids, leaves, and tubers. J. Virol. Methods 91, 37-49. [ Links ]
Offei, S.K., N. Arciniegas, G. Müller, M. Guzmán, L.F. Salazar, and R.H. Coutts. 2004. Molecular variation of Potato Yellow Vein Virus isolates. Arch. Virol. 149, 821-827. [ Links ]
Pruss, G., X. Ge, X.M. Shi, J.C. Carrington, and V.B. Vance. 1997. Plant viral synergism: the potyviral genome encodes a broadrange pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9, 859-868. [ Links ]
Rodríguez-Burgos, P., G. Chaves, L. Franco-Lara, and M. Guzman- Barney. 2009. Low molecular variability of Potato yellow vein virus (PYVV) isolated from Solanum phureja and Solanum tuberosum in Colombia. Phytopathol. 100, S176. [ Links ]
Rodríguez, P., A. Villamil, A. Hernández, M. Guzman-Barney, and W. Cuellar. 2011. Evidencia de coinfección natural de Crinivirus y Potyvirus en aislados colombianos de papa Solanum tuberosum evaluada por secuenciación profunda. Fitopatol. Colomb. 35(suppl.), 60. Memorias XXX Congreso Colombiano y XV Latinoamericano de Fitopatología. Bogota. [ Links ]
Saitou, N. and M. Nei. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406-425. [ Links ]
Salazar, L., G. Müller, M. Querci, J.L. Zapata, and R.A. Owens. 2000. Potato yellow vein virus: its host range, distribution in South America and identification as a Crinivirus transmitted by Trialeurodes vaporariorum. Ann. Appl. Biol. 137, 7-19. [ Links ]
Sánchez de Luque, C., P. Corzo, and O.A. Pérez. 1991. Incidencia de virus en papa y su efecto sobre rendimiento en tres zonas agroecologicas de Colombia. Rev. Latinoam. Papa 4, 36-51. [ Links ]
Schubert, J., V. Fomitcheva, and J. Sztangret-Wisniewska. 2007. Differentiation of Potato virus Y strains using improved sets of diagnostic PCR-primers. J. Virol. Methods 140, 66-74. [ Links ]
Singh, R., J. Valkonen, S. Gray, N. Boonham, R. Jones, C. Kerlan, and J. Schubert. 2008. Discussion paper: the naming of Potato virus Y strains infecting potato. Arch. Virol. 153, 1-13. [ Links ]
Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 101, 11030-11035. [ Links ]
Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA 4: molecular evolutionary genetics analysis (MEGA ) software version 4.0. Mol. Biol. Evol. 24, 1596-1599. [ Links ]
Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTA L W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673-4680. [ Links ]
Untiveros, M., S. Fuentes, and L. Salazar. 2007. Synergistic interaction of Sweet potato chlorotic stunt virus (Crinivirus) with carla-, cucumo-, ipomo-, and potyviruses infecting sweet potato. Plant Dis. 91, 669-676. [ Links ]
Vance, V. 1991. Replication of potato virus X RNA is altered in coinfections with Potato virus Y. Virology 182, 486-494. [ Links ]
Vance, V., H. Berger, J. Carrington, A. Hunt, and X. Shi. 1995. 5' proximal potyviral sequences mediate Potato virus X/potyviral synergistic disease in transgenic tobacco. Virology 206, 583-590. [ Links ]
Vreugdenhil, D. 2007. Potato biology and biotechnology advances and perspectives. Elsevier, Amsterdam. [ Links ]
Wang, J., M. Turina, V. Medina, and B. Falk. 2009. Synergistic interaction between the Potyvirus, Turnip mosaic virus and the Crinivirus, Lettuce infectious yellows virus in plants and protoplasts. Virus Res. 144, 163-170. [ Links ]
Zerbino, D. and E. Birney. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821-829. [ Links ]













