Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Agronomía Colombiana
Print version ISSN 0120-9965
Agron. colomb. vol.33 no.1 Bogotá Jan./Apr. 2015
https://doi.org/10.15446/agron.colomb.v33n1.47988
Doi: 10.15446/agron.colomb.v33n1.47988
1 Oil Palm Biology and Breeding Research Program, Colombian Oil Palm Research Center (Cenipalma). Bogota (Colombia)
2 Department of Biology, Faculty of Sciences, Universidad Nacional de Colombia. Bogota (Colombia). hmromeroa@unal.edu.co
Received for publication: 20 December, 2014. Accepted for publication: 30 March, 2015.
ABSTRACT
Oil palm (Elaeis guineensis Jacq.) fruits are classified by shell thickness into three types: dura, pisifera, and tenera, the last one being the product of a dura × pisifera cross. The palm oil industry relies on the use of high-yield tenera plant material for production; however, it is usually generated with female infertile pisifera, so early identification of this trait is very important to oil production and breeding programs. Recently, the mapping and sequencing of the SHELL gene, which is responsible for endocarp formation in oil palms, made it possible to identify two mutations (type SNP, single nucleotide polymorphism) that affect its function and that are useful to developing molecular markers for predicting shell thickness. The aim of this study was to standardize PCR-based methodologies in order to detect the SNP observed in codon 30 and validate it under our E. guineensis biological collections. We achieved the differentiation of SHELL alleles with both allele specific PCR and CAPS with the restriction enzyme HindIII in homozygous and heterozygous plants that contained the described mutation, and the prediction was correlated with the phenotype observed in oil palm fruits. These methodologies facilitated the discrimination of plants by fruit type in nursery and pre-nursery stages 24 months before production started, thereby reducing the time and area used in oil palm breeding programs.
Key words: SHELL gene, endocarp thickness, fruit type, oil yield, molecular breeding.
RESUMEN
De acuerdo con el grosor del cuesco, los tipos de fruto de palma de aceite (Elaeis guineensis Jacq.) se clasifican en tres: dura, pisifera y tenera; siendo el último la variante heterocigoto de los alelos que generan los dos primeros. La mayor productividad, en términos de cantidad de aceite en el cultivo, se logra al sembrar únicamente materiales tipo tenera, sin embargo estos se generan utilizando como parental el material pisifera, el cual presenta en la mayoría de los casos infertilidad femenina. Recientemente el mapeo genético y secuenciación del gen SHELL, el cual está involucrado en la formación del cuesco en el fruto, permitió determinar que las variantes fenotípicas en el fruto se deben a dos mutaciones tipo SNP (polimorfismo de un solo nucleótido) en este, lo cual permitiría el desarrollo de marcadores moleculares capaces de diferenciar estas mutaciones. El objetivo de este trabajo es estandarizar una metodología basada en PCR, que permita identificar el SNP observado en el codón 30 y validarlo en las colecciones biológicas con las que se trabajó. Mediante PCR alelo específico y CAPS con la enzima de restricción HindIII, se logró identificar la mutación del codón 30 en individuos homocigotos y heterocigotos y el fenotipo predicho concordó perfectamente con el tipo de fruto observado. Estas metodologías permiten la discriminación de plantas de palma de aceite por tipo de fruto en las fases de vivero y previvero, hasta 24 meses antes de que empiece la fase productiva, con la intención de reducir el espacio y el tiempo de los programas de mejoramiento vegetal.
Palabras clave: gen SHELL, grosor de endocarpio, tipo de fruto, rendimiento de aceite, selección asistida por marcadores moleculares.
Introduction
One of the more important traits that define oil yield in oil palms is the thickness of the endocarp, or shell. This economically important fruit trait is controlled by the SHELL gene, which exhibits a co-dominant monogenic inheritance (Beirnaert and Vanderweyen, 1941). SHELL (Sh) encodes a transcriptional activator factor homologous to the gene SEEDSTICK, which is responsible for seed formation and ovule identity in Arabidopsis thaliana (Singh et al., 2013a). A single nucleotide change in codon 28 or 30 impairs the normal DNA binding of shell (sh), leading to a shell-less phenotype (Singh et al., 2013a). Oil palm trees can be classified by SHELL genotype into: dura (Sh/Sh), which is characterized by the production of large fruits with a thick shell and a small proportion of oil-bearing mesocarp, and pisifera (sh/sh), which results in shell-less but mostly female sterile palms (Beirnaert and Vanderweyen, 1941; Obasola, 1973). In addition, hybrids (tenera) between dura and pisifera display a different phenotype, producing fruits with a thinner shell that are smaller and have a larger proportion of oil-bearing mesocarp. The hybrids are therefore associated with a higher oil yield, as compared to dura types. The hybrid vigor observed in tenera palms is associated with a phenomenon of hetero-dimerization, further described by Singh et al. (2013a).
Given the central role that the SHELL gene plays in production, oil palm breeding uses a reciprocal recurrent selection of maternal (dura) and paternal (pisifera) pools. Nonetheless, female infertile pisiferas are the most desirable pollen source for the production of high-quality teneras (Sparnaaij, 1969) although use of female fertile pisiferas is gaining acceptance (Corley and Tinker, 2003). Because infertile pisiferas usually do not produce ripe seeds, the availability of parental donors depends on a chance appearance during genetic segregation in tenera × tenera or tenera × pisifera crosses, requiring large field trials to increase their chance of appearance.
Because oil palm is a perennial species, it is impossible to determine the fruit phenotype until it is reproductively mature (at least 4 years after germination), so the selection of tenera and pisifera is always made in the field and is a current bottleneck for the optimal use of land and financial resources. Here, we propose marker-assisted selection for the SHELL gene based on allelic-specific PCR, which is a rapid and cost-effective technology, would facilitate an effective selection of tenera and pisifera palms in nurseries.
Materials and methods
Single nucleotide polymorphism-SNP identification in studied population and DNA extraction
We studied the SNPs in both codons reported to be responsible for the shell-less fruit phenotype in eight pisifera plants by sequencing exon 1 of SHELL from oil palm breeding populations including Deli, Ekona, Nigeria, La Mé, Papua and AVROS established in the Palmar de la Vizcaina Experimental Field (Cenipalma, Barrancabermeja, Colombia). DNA extraction from plant leaf material was carried out with the DNeasy Plant Mini Kit (Qiagen®, Ref: 69106 Courtaboeuf, France) according to the manufacturer's instructions. Exon 1 SHELL gene amplification was carried out with M13-SHELL primers according to Singh et al. (2013a).
Primer design
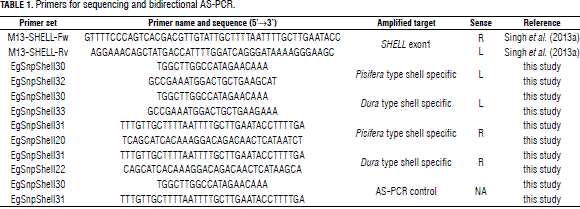
To distinguish between the Sh and sh alleles, four modified allele-specific PCR primers were designed as left or right primers using the web-based software Web SNAPPER (Drenkard et al., 2000), which introduces additional mismatches to enhance the specificity of the primers. The additional mismatch was selected according to the criteria described by Liu et al. (2012), although primer EgSnpShell20 lacks said additional mismatch. Briefly, because the mutation disrupting the shell development arises through an adenine to thymine substitution, the third base closest to the 3'-end of the AS-PCR primers was replaced. Primers EgSnpShell22 and EgSnpShell32 were designed for the sh allele while primers EgSnpShell20 and EgSnpShell33 anneal to the Sh allele. Both the EgSnpShell31 and EgSnpShell30 primers were designed using the primer BLAST web tool (Ye et al., 2012) and function as forward and reverse primers, respectively. All of the primers were checked for primer dimers and hairpin loops both with themselves and all other designed primers using the OligoAnalyzer web tool from IDT SciTools (Owczarzy et al., 2008). Details on the final primer sequence and other related information are listed in Tab. 1.
AS-PCR conditions
PCRs were performed with 25 mL volumes using Taq® polymerase (Thermo Scientific, Waltham, MA). The final concentrations were as follows: 1X Taq buffer, 2.5 mM MgCl2, 0.32 mM dNTP mixture, 0.5 mM for each primer, 0.5 U Taq and 50 ng gDNA. All reactions were performed using a C1000 Thermal Cycler (Bio-Rad, Hercules, CA) with an initial denaturing cycle of 3 min at 95°C, 25 cycles of 30 s at 95°C, 1 min at 60.7°C, 1 min at 72°C, and a final extension cycle of 5 min at 72°C. The PCR products were visualized using EZ-Vision DNA Dye as a loading buffer (Amresco®, Solon, OH) in 2% agarose gels.
CAPS analysis
EgSnpShell30 and EgSnpShell31 primers were used for the initial amplification step to obtain a fragment of SHELL. The employed PCR protocol was modified using a touchdown gradient approach, decreasing the annealing temperature by 0.5°C each cycle, from 64 to 59°C, by running 10 cycles; then, the annealing was maintained at 59°C for the next 25 cycles. The primer concentration was 1 mM and the PCR products were visualized with 2.0% agarose gel electrophoresis. Then, 20 mL of the PCR product were digested with 15 units of HindIII restriction enzyme (Thermo Scientific, Waltham, MA) with an appropriated buffer at 37°C for 18 h. The digestion products were visualized with 2.3% agarose gel electrophoresis.
Results
There was a single nucleotide difference between adenine (A) and thymine (T), encoding an amino acid substitution of lysine (AAA) with asparagine (AAT) at residue 30 of the SHELL protein. However, no mutation was detected at codon 28 in the tested population. Based on this nucleotide mutation, allele-specific primers were designed.
Primers EgSnpShell30 and EgSnpShell31 amplify a 446 bp fragment containing the mutation. As all plants should show this fragment, it can serve as an internal positive control. Primers annealing to the Sh and sh alleles specifically resulted in two other amplicons of 310 and 191 bp, respectively (Fig. 1). Primers were first tested in single reactions and all of them were able to discriminate between dura and pisifera DNA as predicted by their initial design.
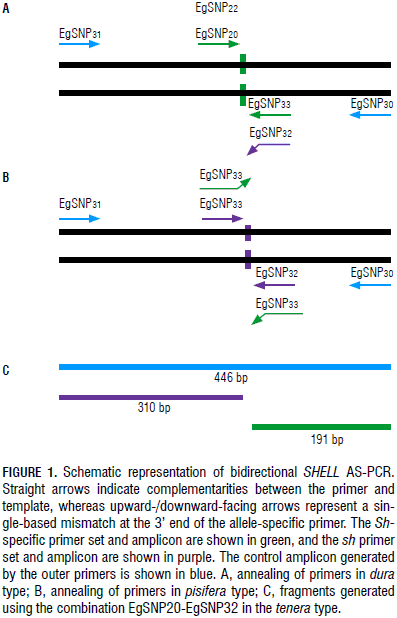
The primers designed as left or right primers were combined in order to differentiate both alleles in a single reaction (Fig. 1). The EgSnpShell22-EgSnpShell33 combination was unsuccessful in amplifying the allelic bands using DNA from dura or pisifera plants as templates, probably due to competition between the primers for PCR amplification; therefore, the EgSnpShell20-EgSnpShell32 combination was selected for further optimization in tenera (data not shown).
Inclusion of multiple allele-specific primer sets in a single reaction tube requires the consideration of multiple experimental factors (Henegariu et al., 1997). We tested three critical parameters whose interactions are most likely to affect the outcome of a bidirectional AS-PCR (Liu et al., 1997): primer set, dNTPs and MgCl2. The concentration of the primers was optimized by testing all possible combinations of concentrations of all primers at 0.5, 1.0 and 2.0 mM using the standard amplification conditions. Genomic DNA from tenera was added and each reaction was tested in a temperature gradient in order to assess the optimal annealing temperature. Standard MgCl2 curves were also tested.
The optimal conditions for zygosity discrimination are those that amplify each allele-specific amplicon at similar efficiencies as measured by band intensity from the gel. In this study, the maximum specificity and product yield were achieved with 2.5 mM MgCl2, and the excess of the forward primer (2:1:1:1 ratio) at 60.7°C gave optimal results, yielding products with similar efficiency. Those conditions were set as the optimized discriminating conditions.
The optimal nature of the discriminating conditions was confirmed unambiguously in a single blind trial of 38 samples of different zygosity. The segregating material for the single blind trial was obtained from the Angola Germplasm Collection of the Palmar de la Vizcaina Experimental Field (Barrancabermeja, Colombia). This collection is composed of seeds from open-pollinated natural populations of dura and tenera palms (Arias et al., 2013). A total of 38 palms, including 15 dura, 13 pisifera, and 10 tenera palms, were tested. The samples were classified based on the band profile only and the results were correlated with the fruit phenotype of each sample (Fig. 2).
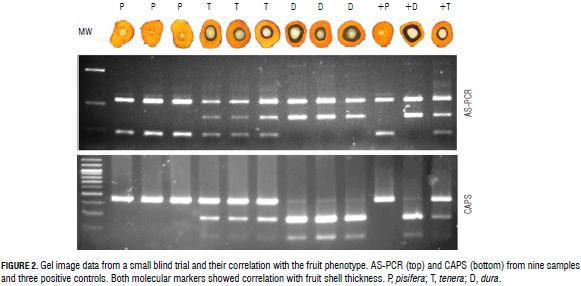
Further validation of this technique was done by identifying a restriction enzyme, which includes the SNP observed in its recognition site. We found that the enzyme HindIII has a recognition site in the Sh allele that is absent in the sh allele due to the A to T change observed in codon 30, so this could be used as a cleaved amplified polymorphic sequence (CAPS) marker for easy and highly reproducible detection of these alleles in both homozygous and heterozygous plants. All samples tested with CAPS confirmed both phenotypic and genotypic analysis with AS-PCR (Fig. 2), corroborating the effectiveness of either AS-PCR or CAPS as a useful tool for discrimination of oil palm plants at the nursery stage if the SNP of SHELL is in codon 30.
Discussion
No discrepancies where found between our results and the observed fruit types, confirming that the primers or the restriction enzyme were capable of effective discrimination. However, a varied degree of shell thickness was observed in the dura and tenera samples, indicating that there is a thickness-related polygenic variation. Because these techniques are gel based, they are cost effective and can be easily implemented in any laboratory. Additionally, although gel-based assays are not usually suitable for large numbers of samples (Bassam et al., 1996), the different pattern for each genotype facilitates the high-throughput application of this technique, allowing for fast and highly reproducible analysis.
Despite obtaining 100% phenotype - genotype correlation in our experiments, there are other SNPs that may affect this trait, including the codon 28 SNP reported by Singh et al. (2013a). According to patent number US 20130247249A1, there is one genomic region of 3.4 Mb that contains the SHELL gene, where more than 8,200 SNPs had been detected, and that can be used as a molecular marker for fruit type prediction with less accuracy (Singh et al., 2013b). In previous studies, markers, such as AFLP and RAPD, were linked to this trait, but they were too far from the SHELL gene and it was impossible to apply these methodologies in oil palm breeding programs (Moretzsohn et al., 2000; Billotte et al., 2001; Billotte et al., 2005). More recently, Gan (2014) identified a set of closely-linked shell-thickness markers through saturation of the Sh region with DArTSeq markers, as well as map integration around the Sh regions in two F2 mapping populations. In addition, this study identified 32 SNP and DArT markers mapped within a 5 cM flanking region of the Sh gene. These markers could be valuable as a molecular screening tool for fruit form determination, but, at this time, there is not a publication based on PCR protocols to identify the fruit type.
Discrimination of SHELL alleles by means of molecular markers may benefit oil palm breeders and seed producers at several levels. For example, for the certification of high-yielding seed production by selective breeding of D×P crosses, particularly on plantations that lack stringent quality control and/or where natural pollination with small quantities of tenera or dura pollen can occur, the selection of true female sterile pisifera parentals and selection without recurring to the progeny of elite tenera from T×T or T×P crosses can be used.
The oil palm industry will continue to grow due to the high global demand for its derivatives and its economic impact on countries such as Indonesia, Malaysia, Thailand, Nigeria, Ecuador and Colombia. However, one of the main challenges for the oil palm industry today is improving crop yield using limited land resources in order to maintain equilibrium with natural ecosystems. The results of the present study have demonstrated the capacity of a bi-directional AS-PCR to detect the genetic variation of the SHELL gene, showing how a simple molecular marker can be successfully used to diagnose an oil palm characteristic in the nursery stage that was previously only observable at the production stage, favoring the selection of desirable high-yielding plant material or donor pollen and avoiding unnecessary land consumption in large field trials.
Acknowledgements
This work was funded by the Oil Palm Development Fund (FFP) managed by Fedepalma.
Literature cited
Arias, D., M. González, F. Prada, E. Restrepo, and H. Romero. 2013. Morpho-agronomic and molecular characterisation of oil palm Elaeis guineensis Jacq. material from Angola. Tree Genet. Genomes 9, 1283-1294. Doi: 10.1007/s11295-013-0637-5 [ Links ]
Bassam, B.J., T. Allen, S. Flood, J. Stevens, P. Wyatt, and K.J. Livak. 1996. Nucleic acid sequence detection systems: revolutionary automation for monitoring and reporting PCR products. Aus. Biotech. 6, 285-294. [ Links ]
Billotte, N., L. Frances, P. Amblard, T. Durand-Gasselin, J.L. Noyer, and B. Courtois. 2001. Search for AFLP and microsatellite molecular markers of the SH gene in oil palm (Elaeis guineensis Jacq.) by bulk segregant analysis (BSA) and by genetic mapping. pp. 442-445. In: Proc. 2001 PIPOC Int. Palm Oil Cong. Cutting-Edge Technologies for Sustained Competitiveness. Kuala Lumpur. [ Links ]
Billotte, N., N. Marseillac, A.-M. Risterucci, B. Adon, P. Brottier, F.-C. Baurens, R. Singh, A. Herrán, H. Asmady, C. Billot, P. Amblard, T. Durand-Gasselin, B. Courtois, D. Asmono, S.C. Cheah, W. Rohde, E. Ritter, and A. Charrier. 2005. Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 110, 754-765. Doi: 10.1007/s00122-004-1901-8 [ Links ]
Beirnaert, A. and R. Vanderweyen. 1941. Contribution à l'étude genetique et biometrique des variétés d' Elaeis guineensis Jacquin. Institut National Pour l'étude Agronomique du Congo Belge (INEAC), Brussels. [ Links ]
Corley, R.H.V. and P.B. Tinker. 2003. The oil palm. 4th ed. Blackwell Science, Oxford, UK. Doi: 10.1002/9780470750971 [ Links ]
Drenkard, E., B.G. Richter, S. Rozen, L.M. Stutius, N.A. Angell, M. Mindrinos, R.J. Cho, P.J. Oefner, R.W. Davis, and F.M. Ausubel. 2000. A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 124, 1483-1492. Doi: 10.1104/pp.124.4.1483 [ Links ]
Gan, S.T. 2014. The development and application of molecular markers for linkage mapping and quantitative trait loci analysis of important agronomic traits in oil palm (Elaeis guineensis Jacq.). PhD thesis. Malaysia Campus, University of Nottingham. Semenyih, Malaysia. [ Links ]
Henegariu, O., N.A. Heerema, S.R. Dlouhy, G.H. Vance, and P.H. Vogt. 1997. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques 23, 504-511. [ Links ]
Liu, J., S. Huang, M. Sun, S. Liu, Y. Liu, W. Wang, X. Zhang, H. Wang, and W. Hua. 2012. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 8, 34. Doi: 10.1186/1746-4811-8-34 [ Links ]
Liu, Q., E.C. Thorland, J.A. Heit, and S.S. Sommer. 1997. Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res. 7, 389-398. Doi: 10.1101/gr.7.4.389 [ Links ]
Moretzsohn, M.C., C.D.M. Nunes, M.E. Ferreira, and D. Grattapaglia. 2000. RAPD linkage mapping of the shell thickness locus in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 100, 63-70. Doi: 10.1007/s001220050009 [ Links ]
Obasola, C.O. 1973. Female sterility and fertility in variety pisifera and shell thickness in progenies of dura x pisifera crosses of the oil palm (Elaeis guineensis Jacq.). I. categories of variety pisifera. J. Niger. Inst. Oil Palm Res. 5, 37-41. [ Links ]
Owczarzy, R., A.V. Tataurov, Y. Wu, J.A. Manthey, K.A. McQuisten, H.G. Almabrazi, K.F. Pedersen, Y. Lin, J. Garretson, N.O. McEntaggart, C.A. Sailor, R.B. Dawson, and A.S. Peek. 2008. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucl. Acids Res. 36(suppl. 2), W163-W169. Doi: 10.1093/nar/gkn198 [ Links ]
Singh, R., E.-T.L. Low, L.C.-L. Ooi, M. Ong-Abdullah, N.-C. Ting, J. Nagappan, R. Nookiah, M.D. Amiruddin, R. Rosli, M.A.A. Manaf, K.-L. Chan, M.A. Halim, N. Azizi, N. Lakey, S.W. Smith, M.A. Budiman, M. Hogan, B. Bacher, A. Van Brunt, C. Wang, J.M. Ordway, R. Sambanthamurthi, and R.A. Martienssen. 2013a. The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 500, 340-344. Doi: 10.1038/nature12356 [ Links ]
Singh, R., E.-T.L. Low, L.C.-L. Ooi, M. Ong-Abdullah, R. Nookiah, R. Sambanthamurthi, S.W. Smith, N.D. Lakey, R. Martienssen, J.M. Ordway, and M. Hogan. 2013b. Gene controlling shell phenotype in palm. Patent Application Publication US 2013/0247249 A1. [ Links ]
Sparnaaij, L.D. 1969. Oil palm (Elaeis guineensis Jacq.). pp. 339-387. In: Ferwerda, F.P. and F. Wit (eds.). Outline of perennial crop breeding in the tropics. Miscelaneous Papers in Agriculture Vol. 4. H Veenman & Zonen NV, Wageningen, The Netherlands. [ Links ]
Ye, J., G. Coulouris, I. Zaretskaya, I. Cutcutache, S. Rozen, and T.L. Madden. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. Doi: 10.1186/1471-2105-13-134 [ Links ]














