Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Agronomía Colombiana
versão impressa ISSN 0120-9965
Agron. colomb. vol.34 no.1 Bogotá jan./abr. 2016
https://doi.org/10.15446/agron.colomb.v34n1.52960
Doi: 10.15446/agron.colomb.v34n1.52960
1 Department of Agronomy, Faculty of Agricultural Sciences, Universidad Nacional de Colombia. Bogota (Colombia). michacons@unal.edu.co
2 Tibaitata Research Center, Corporacion Colombiana de Investigacion Agropecuaria (Corpoica). Mosquera (Colombia)
Received for publication: 8 September, 2015. Accepted for publication: 28 March, 2016.
ABSTRACT
The cape gooseberry (Physalis peruviana L.), a fruit species cultivated in the Andes, is one of the major fruit exports of Colombia. We hypothesized that the Andean cordilleras in Colombia play a role in structuring the genetic diversity of this crop. For addressing this hypothesis, a set of 85 Colombian cape gooseberry accessions from different departments and cordilleras was analyzed by means of 15 SSR markers. AMOVA, clustering and Bayesian analyses were applied. The results showed the presence of two major groups related to geography: one consisting of cultivated and non-cultivated accessions from the eastern Andes (Norte de Santander, Santander, Boyaca and Cundinamarca) and the other one consisting of cultivated and non-cultivated accessions from the central and western Andes (Antioquia, Caldas, Cauca and Nariño). The genetic relationships between the accessions suggested that the movement of cape gooseberry seeds may be more frequent between neighboring regions, thus explaining the existence of these two major groups. The results also showed lower levels of genetic diversity in this sample (HE=0.223), as compared to other Physalis species and other studies on the cape gooseberry that used different molecular markers. It is recommended that future evaluation studies include both cultivated and non-cultivated genotypes from the two major groups detected in this study in order to better represent the genetic diversity available in this crop.
Key words: germplasm, cultivars, Andean region, genetic diversity as resource, genetic distance, molecular markers.
RESUMEN
La uchuva (Physalis peruviana L.), una de las frutas más importantes para la exportación en Colombia, es cultivada en las tres cordilleras de los Andes. Este patrón de distribución geográfica nos lleva a formular que las cordilleras de los Andes juegan un papel importante en la estructura genética de este cultivo. Para abordar esta hipótesis, la estructura genética en un conjunto de 85 accesiones cultivadas y no cultivadas de uchuva se estudió por medio de 15 loci SSR y se aplicaron análisis de AMOVA, agrupamiento y métodos Bayesianos. Los resultados muestran la existencia de dos grupos de accesiones cultivadas y no cultivadas: uno ubicado en la cordillera oriental de los Andes (Norte de Santander, Santander, Boyacá y Cundinamarca) y el otro en la cordillera central y occidental de los Andes (Antioquia, Caldas, Cauca y Nariño). Las relaciones genéticas entre accesiones sugieren que las semillas de uchuva son transportadas preferiblemente entre regiones cercanas, generando así la estructura genética observada. Se observaron niveles de diversidad genética más bajos (HE=0,223) en la muestra analizada que en otras especies de Physalis o en otros estudios de uchuva con diferentes marcadores moleculares. Se recomienda que futuros estudios de evaluación de la uchuva incluyan accesiones cultivadas y no cultivadas que representen los dos grupos observados en la presente investigación con el fin de representar mejor la diversidad genética disponible en esta especie.
Palabras clave: germoplasma, cultivares, Adean region, diversidad genética como recurso, distancia genética, marcadores moleculares.
Introduction
The cape gooseberry (Physalis peruviana L.), a species of the Solanaceae family and native to the Andes of South America (Cailes, 1952; Legge, 1974; Council, 1989), is an economically important fruit crop in Colombia. Two important life history traits that can affect genetic diversity, namely the ploidy level and mating system, have been previously studied in this species. In 1951, Menzel reported a chromosome number of 2n=48 for P. peruviana (Menzel, 1951), but recent studies have demonstrated polymorphism in the chromosome number. Rodríguez and Bueno (2006) observed three different chromosome numbers in five individuals that were analyzed; 2n=24 for wild genotypes, 2n=32 for the Colombia ecotype and 2n=48 for the Kenya ecotype. Lagos et al. (2008) investigated the mating system in the cape gooseberry under greenhouse conditions and observed a mixed mating system with 54% outcrossing and the absence of mechanisms promoting self-incompatibility.
In Colombia, the cape gooseberry grows as a wild plant in the Andes at elevations ranging from1,500 to 3,000 m and as a cultivated plant between 2,400 and 3,400 m, at average temperatures between 8 and 14°C (Fischer and Almanza, 1993; Fischer, 2000). According to records from the Colombian government, during the years 1995-2000, the cape gooseberry production in Colombia was concentrated in the departments of Cundinamarca and Tolima and starting in the year 2000, its cultivation spread to Boyaca, Antioquia, Valle del Cauca, Cauca, Nariño and Norte de Santander (Agronet, 2015). In the year 2013, a total of 880 ha were cultivated with a production of 12,873 t and a yield of 14.6 t ha-1 (Agronet, 2015). With the expansion of the cultivated area, many agronomical limitations have arisen; among the most limiting factors is the disease caused by the soilborne fungus Fusarium oxysporum (González and Barrero, 2011).
More knowledge and conservation of the native cape gooseberry germplasm is needed to overcome the current agronomical problems. The National Germplasm Bank of Colombia, managed by the Corporacion Colombiana de Investigacion Agropecuaria (Corpoica), holds a collection of 57 cape gooseberry accessions, of which 48 are native to Colombia, mainly from Antioquia, Boyaca, Caldas, Cundinamarca and Nariño, and nine are from other countries (Valencia et al., 2010). The Faculty of Agricultural Sciences of the Universidad Nacional de Colombia (UNAL) in Bogota holds a collection of 54 accessions obtained from cultivated fields and from plants growing spontaneously (wild or feral) in Cundinamarca, Boyaca, Santander and Norte de Santander. Likewise, UNAL in Palmira holds a collection of nearly 200 accessions from southwestern Colombia. The Universidad de Nariño (UDENAR) holds a collection of about 50 accessions of cape gooseberry, mainly from Nariño and Cauca.
These cape gooseberry collections have been partly evaluated for morpho-agronomic variables and resistance to F. oxysporum, a key trait to improve the production of cape gooseberries in Colombia. UDENAR and UNAL in Palmira have pioneered the study of the genetic and agro-morphological diversity of cape gooseberries from southwestern Colombia (Criollo et al., 2001a; Criollo et al., 2001b; Lagos et al., 2001; Bonilla and Espinosa, 2003; Bonilla et al., 2008) and UNAL in Bogota and Corpoica have mainly studied accessions from Antioquia, Cundinamarca, Boyaca, Santander and Norte de Santander (Herrera et al., 2011; Herrera et al., 2012; Enciso-Rodríguez et al., 2013; Berdugo et al., 2015; Garzón-Martínezet al., 2015). These studies have shown that cultivated accessions are superior in terms of yield and fruit weight, while wild accessions are superior in terms of number of fruits per plant, absence of fruit cracking and high concentration of total soluble solids (Herrera et al., 2011; Herrera et al., 2012). Additionally, wild accessions from certain geographic regions exhibit a tolerance response to F. oxysporum under specific experiment conditions, specially one from the department of Nariño (Enciso-Rodríguez et al., 2013).
In spite of being an economically important species, only recently have some studies documented the genetic diversity of the Colombian cape gooseberry using molecular markers. Among the most useful molecular markers for the study of plant germplasm are the microsatellites or SSR (short sequence repeats), which are tandem repeats of DNA motifs that consist of one to six nucleotides (Kalia et al., 2011). Two main properties have made SSR the markers of choice for population genetic studies in many organisms: they are highly polymorphic and codominant. Among the studied organisms are Andean crops of the genera Solanum, Rubus and Passiflora, among others (Ortiz et al., 2012; Bushakra et al., 2015; Juyóet al., 2015). For the cape gooseberry, Simbaqueba et al. (2011) developed a set of SSR markers from the cape gooseberry leaf transcriptome sequenced by Garzón-Martínezet al. (2012). From these sequences, SSR loci were detected in coding and non-coding regions with a bioinformatics approach and then PCR primer pairs were designed for 162 SSR loci located in non-coding regions. These 162 loci were evaluated in seven cape gooseberry accessions and the species Physalis floridana Rydb., and of these loci, 30 were identified as polymorphic.
Other studies have explored the genetic diversity of the Colombian cape gooseberry with molecular markers of different natures. Enciso-Rodríguez et al. (2013) sequenced 74 genes related to plant immunity in six Physalis sp. accessions with variable responses to F. oxysporum and detected one single-nucleotide polymorphism (SNP) candidate related to resistance. Osorio-Guarínet al. (2016) analyzed the genetic diversity of a collection of around 100 cape gooseberry accessions from different departments and biological statuses by means of about 5,000 SNP markers and concluded that the genetic structure was mainly associated with the state of cultivation (wild and cultivated) and not to geography. On the other hand, Garzón-Martínezet al. (2015) studied the genetic diversity and population structure of 47 accessions of P. peruviana, mainly from the departments of Cundinamarca, Boyaca and Antioquia, and 13 related taxa. These authors analyzed 642 SNP markers and 24 InDels (insertion or deletion) loci and concluded that the genetic diversity was organized into two main groups, corresponding to the cultivation status (wild and cultivated) and not to the geographic origin.
As mentioned before, wild and cultivated cape gooseberries in Colombia are found in the Andean region, a region that is separated into three mountain chains or cordilleras (western, central and eastern Andes). Therefore, we hypothesized that the genetic diversity in this species is influenced by the topography of the Colombian Andes. Previous studies have reported that the genetic structure is related mainly to the state of cultivation and not to geography. In this research, we used the SSR loci reported by Simbaqueba et al. (2011) to further investigate the role of the Colombian Andes and the geographic origin of the accessions in determining the genetic structure of the cape gooseberry. We also took advantage of morpho-agronomical data generated in previous studies in some of these accessions (Herrera et al., 2011; Herrera et al., 2012) to carry out association tests between SSR polymorphisms and variables related to yield and fruit quality. These results will enhance our understanding of how diverse the cape gooseberry germplasm of Colombia is and will give insights on how future collection, conservation and improvement activities should be conducted to better exploit the genetic diversity present in this species.
Materials and methods
Plant material
A total of 345 individuals, distributed into 32 cultivated and 53 non-cultivated accessions, was analyzed, with an average of four individuals per accession. Of these, 54 accessions were obtained from UNAL in Bogota, seven accessions from Corpoica in Mosquera (Cundinamarca), 18 accessions from Corpoica Rionegro (Antioquia) and six accessions from Universidad de Nariño. The distribution of these accessions by department and by Andean cordillera can be seen in Tab. 1. Cultivated accessions are those collected in cultivated fields for commercialization purposes. Non-cultivated accessions are those collected outside cultivated fields from plants growing spontaneously (either wild or feral), mostly in disturbed areas near gardens and roads. We prefer to use the term non-cultivated to include wild and feral populations of the cape gooseberry.
Molecular analyses
Genomic DNA was extracted for each individual from young leaves using the protocol reported by Vega-Vela and Chacón-Sánchez (2011). A total of 15 SSR loci that proved to be polymorphic in a previous study (Simbaqueba et al., 2011) were analyzed in the cape gooseberries. The SSR loci were: SSR1, SSR2, SSR10, SSR11, SSR15, SSR54, SSR72, SSR77, SSR112, SSR118, SSR121, SSR123, SSR126, SSR138 and SSR146. Information about primers, melting temperature and conditions for PCR amplification can be found in Simbaqueba et al. (2011). The forward primer of each locus was fluorescently labeled at the 5' end. After amplification, PCR products were separated in an ABI PRISM® 3500 (Applied Biosystems, Foster City, CA) through services provided by the Institute of Genetics at UNAL in Bogota. Allele calling and genotyping were carried out with the program GeneMapper® v. 4.0 (Applied Biosystems, 2006).
Genetic structure analyses
To quantify the genetic diversity of the accessions according to their geographic origin and biological status (cultivated and non-cultivated), we calculated the number of alleles per locus (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (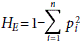 ) and fixation index (F=(HE-HO)/HE) with the program GenAlex v. 6.5 (Peakall and Smouse, 2012). For each locus, the polymorphism information content (PIC) was also calculated.
) and fixation index (F=(HE-HO)/HE) with the program GenAlex v. 6.5 (Peakall and Smouse, 2012). For each locus, the polymorphism information content (PIC) was also calculated.
In order to evaluate the genetic relationships between the accessions by geographic origin and biological status, two matrixes of genetic distances were calculated. Nei's genetic distance assumes the mutational model of infinite alleles (Nei, 1987) and the chord distance (Cavalli-Sforza and Edwards, 1967) does not assume any specific mutational model. From these distances, neighbor-joining topologies were generated with the PHYLIP package v. 3.5c (Felsenstein, 1993) and the topologies were visualized with the software FigTree (Rambaut and Drummond, 2014).
The genetic structure was investigated by means of analysis of molecular variance (AMOVA) (Excoffier et al., 1992) with 1,000 permutations as implemented in the program GenAlEX v. 6.5 (Peakall and Smouse, 2012). For this purpose, we used the state of cultivation (cultivated and non-cultivated), department of origin and Andean cordillera (western, central and eastern Andes) as sources of variation. The genetic structure was also investigated by means of a Bayesian approach implemented in the software Structure (Pritchard et al., 2000) and by means of a principal coordinate analysis (PCO) implemented in GenAlEX v. 6.5 (Peakall and Smouse, 2012). For the Bayesian approach, individuals were assigned to a number of K populations and Q-matrixes of global ancestry were obtained. In this analysis, the K from 2 to 6 was analyzed in a total of 20 simulations with a burnin period of one million and one million MCMC (Markov chain Monte Carlo) steps after burnin. The program CLUMPP (Jakobsson and Rosenberg, 2007) was used to obtain a single Q-matrix for each K and from all of the 20 independent simulations. The optimum K was chosen according to Evanno et al. (2005) using the Structure Harvester program (Earl and VonHoldt, 2012). Bar plots for the ancestry coefficients of individuals were drawn using the Distruct software (Rosenberg, 2007). The admixture and correlated allele frequency models were used because they are more appropriate for outcrossing species such as the cape gooseberry (Lagos et al., 2008). For the PCO analysis, the individuals were grouped according to their geographic origin and biological status and a matrix of genetic distance was calculated with the program GenAIEX v. 6.5 (Peakall and Smouse, 2012).
Morpho-agronomical traits and association analyses
Herrera et al. (2011, 2012) carried out a morpho-agronomical evaluation of a collection of 54 cape gooseberry accessions held at the UNAL, Bogota, with 23 quantitative variables related to yield and fruit quality, among them the most important variables were: yield, total soluble solids, percentage of fruit cracking, fresh fruit weight with the calyx, fresh fruit weight, dry fruit weight with the calyx, dry fruit weight, fruit volume, pH and maturity index. In this study, association tests were carried out between the SSR allele diversity and these ten phenotypic variables for a sample of 187 individuals from which these data were available. For this, mixed linear models implemented in the software GAPIT (Lipka et al., 2012) were used, controlling for the population structure and genetic relatedness among the individuals by incorporating a PCA and a kinship matrix also calculated in GAPIT (Lipka et al., 2012). Box-Cox transformations of variables were done with the appropriate lambda values with the software R (Team, 2014) to reach the optimal fit against the normal distribution. P-values were corrected using the false discovery rate (FDR) criterion.
Results
Polymorphisms in SSR loci
A total of 50 alleles was observed in the whole sample, between two and five alleles per locus and an average number of alleles per locus of 3.059 (Tab. 2). It can be seen that the observed allele sizes were close to the expected allele sizes, that many alleles were rare (frequency <5%) and that many rare alleles were unique to either cultivated or non-cultivated accessions. The most polymorphic SSR locus was SSR15 with a PIC of 0.568, the remaining SSR loci showed PIC values between 0.043-0.067, 0.101-0.132 and 0.317-0.344. The expected heterozygosity ranged from 0.026 for locus SSR121 and SSR10 to 0.641 for locus SSR15, with an average expected heterozygosity of 0.223. The inbreeding coefficients (F) for these loci were mostly negative, indicating an excess of heterozygotes, as can be expected for a self-compatible but outcrossing species (Lagos et al., 2008).
Genetic structure
When comparing the accessions from different biological statuses (Tab. 3), we saw that the cultivated and non-cultivated accessions were very similar in terms of different indexes related to genetic diversity (Na, Ne, Ho, HE and F). The genetic differentiation among the cultivated and non-cultivated accessions as measured by the AMOVA was small but significant (FST = 0.058, P = 0.001).
Table 3 also shows the genetic diversity values estimated according to the department of origin. The expected heterozygosity (HE) values ranged from 0.173 for Antioquia to 0.229 for Boyaca and Norte de Santander. The genetic differentiation among the departments (FST = 0.106, P= 0.001), as indicated by the AMOVA, was significant and larger than between the biological statuses.
In regards to the Andean cordilleras, the HE values ranged from 0.137 for the central Andes to 0.192 for the eastern Andes. The level of differentiation between the Andean cordilleras (FST = 0.174, P = 0.001) was significant and larger than between the departments and biological statuses.
The genetic structure was also investigated by means of genetic distances, PCO and structure analyses. The average Nei's genetic distance among the accessions was 0.13, with a minimum value of 0 and a maximum value of 0.503. Figure 1 shows a histogram of genetic distance values among the analyzed accessions. Most of the pairwise comparisons showed genetic distance values between 0.1 and 0.15. The genetic distance was also calculated for the accessions from different departments and biological statuses (13 different groups). The department that showed the highest average genetic distance was Cauca with 0.19, much higher than the average of 0.13. The pairwise comparison between the accessions that showed the largest genetic distance (0.50) was between one accession from Cauca (non-cultivated) and one accession from Norte de Santander (non-cultivated), two departments that are at the two extremes of the analyzed geographical range.
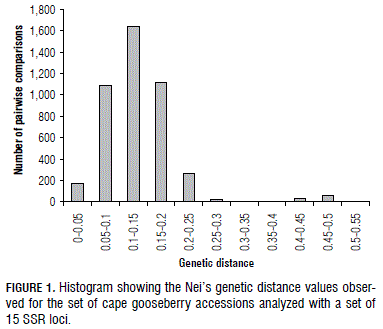
Figure 2 shows the genetic relationships among the accessions according to the department of origin and biological status by means of a neighbor joining (NJ) topology based on Nei's genetic distances (similar results were obtained with the Chord distance). It can be seen that, on the one hand, cultivated and non-cultivated accessions from the eastern Andes tended to cluster together (group 1) and, on the other hand, cultivated and non-cultivated accessions from the central and western Andes tended to cluster together (group 2). The accession from Cauca (non-cultivated) appeared as the most distant accession.
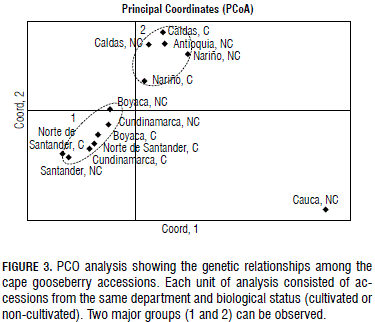
The three axes of PCO explained 57.36%, 24.04% and 10.20% of variation, respectively, for a cumulative 91.60%. The PCO analysis also suggested the existence of two main groups of accessions (Fig. 3), similar to the NJ topology. The non-cultivated accession from Cauca was separated from the two groups, a pattern also observed in the NJ topology.
For the Bayesian analysis in the structure, 13 groups were defined according to the department of origin and biological status (see legend of Fig. 4). The optimum K was 4 according to the method proposed by Evanno et al. (2005). Figure 4 shows the plot of ancestry coefficients from K 2 to 4 for each of the 13 groups. Figure 5 shows the ancestry coefficients for each of the 13 groups from each of the four populations defined by the structure (K = 4). It can be seen in Fig. 5 that the ancestry coefficients of the departments from the eastern Andes (group 1) were different from those at the central and western Andes (group 2). The differentiation between these two groups as measured by the AMOVA (FST = 0.181, P = 0.001) was significant.
Association analyses among the SSR alleles and morpho-agronomical variables
No associations were detected among the SSR alleles and the ten analyzed phenotypic variables. For comparison purposes, association analyses were carried out with raw data and transformed data. With the raw data, two alleles in the locus SSR2 were associated with the fruit weight, fruit weight with the calyx and fruit volume, and one allele in the locus SSR54 was associated with the maturity index and pH, but these associations did not hold when the transformed data were used.
Discussion
Documenting genetic diversity patterns in an economically important species is key for future applications and conservation of the germplasm. For example, when selecting pa- rental genotypes for breeding programs or QTL mapping, it is very useful to know how distantly related the selected genotypes are. Also, for ex situ conservation purposes and for germplasm collection activities, it is important to know how the genetic diversity available in situ is structured. Hopefully, the information generated in this study will help future investigators make informed decisions about the use of the cape gooseberry germplasm.
The main purpose of this study was to assess the genetic structure of the Colombian cape gooseberries from different geographic regions (departments and Andean cordilleras). We start this section by discussing the polymorphisms observed in the SSR loci, then we examine the levels of genetic diversity found in P. peruviana and we end our discussion by comparing the role of the biological status and geographic origin in the organization of genetic diversity.
Polymorphism of SSR loci
In this study, a set of 15 microsatellite markers were used to assess the genetic diversity patterns in cape gooseberries from different geographic regions in Colombia. In this study, PIC values for the 15 SSR loci ranged from 0.043 to 0.568, with an average of 0.215, and only six loci showed moderately high PIC values ranging from 0.317 to 0.568. These PIC values are similar to those reported by Garzón- Martínezet al. (2015) (PIC values from 0.094 and 0.663) who analyzed a set of 47 P. peruviana accessions and 13 related taxa with 24 InDel markers derived from single copy genes in the nuclear genome. Although the SSR loci used here proved to be useful for genetic diversity studies on this species, further studies should screen a larger number of SSR loci in order to identify markers with high PIC values (PIC of 0.6 and above) that would be useful for germplasm identification purposes (for example, variety identification).
Genetic diversity of cape gooseberries in Colombia
When interpreting the genetic diversity values obtained in one study, it is misleading to refer to those values in absolute terms and, therefore, it is better to make comparisons with previous studies on the same or related species with similar molecular markers. By doing so, we found that the Colombian cape gooseberry accessions analyzed here contained slightly lower or similar numbers of alleles per locus (Na = 3.059) than other Physalis species. Wei et al. (2012) studied a collection of 22 Physalis philadelphica Lam. (or tomatillo) accessions, mainly from Mexico, and 14 accessions of other Physalis species, including three of P. peruviana, by means of 25 SSR loci developed by Simbaqueba et al. (2011). For these 25 SSR loci, the authors reported Na values of 3.3 for P. philadelphica and 4.44 for the other Physalis species. When comparing the Nei's genetic distances, we observed that, while in cape gooseberry the average Nei's genetic distance was 0.13, for the tomatillo, the average Nei's genetic distance was almost double (0.22) (Wei et al., 2012).
In other types of comparisons, we found that the expected heterozygosity (HE = 0.223) estimated in the present study was lower than in previous studies involving the Colombian cape gooseberry. For example, Osorio-Guarínet al. (2016) studied a set of 100 wild and cultivated cape gooseberry accessions from different regions in Colombia with more than 5.000 SNP markers and reported a HE value of 0.647, almost three times the one reported here. This strong discrepancy may be explained by the kind of marker used in each study, the differences in accessions used, and the amount of loci analyzed, with the study by Osorio-Guarínet al. (2016) using a more complete genomic sampling. On the other hand, Garzón-Martínezet al. (2015) reported HE values for the Colombian cape gooseberry of 0.30 for InDel markers and 0.41 for SNP markers, again HE values higher than the one reported here.
In summary, we can see that the magnitude of the genetic diversity estimators may differ greatly among studies and this depends on many factors, such as sample size, origin accessions and molecular markers that are used. So far, the most complete genomic sampling of the Colombian cape gooseberry has been the one reported by Osorio-Guarínet al. (2016), where higher values of genetic diversity were reported (HE = 0.647). This suggests that the potential of the Colombian cape gooseberry to harbor alleles that are useful for a variety of breeding purposes is promising and still to be exploited.
Genetic structure of the Colombian cape gooseberry
The genetic structure of the Colombian cape gooseberry by biological status and geography was explored in this study. The genetic relationships among the different geographical regions and biological statuses shown in Fig. 2 and the clustering patterns shown by the PCO and structure analyses suggest that the Colombian cape gooseberry could be clustered into two groups, mainly by geography. Group 1 contained accessions from the eastern Andes (from Cundinamarca to Norte de Santander) and group 2 contained accessions from the central and western Andes (from Antioquia to Nariño). These results suggest that the Andean cordilleras play an important role in structuring the genetic diversity in this species and that cape gooseberry seeds may be preferentially exchanged between neighboring regions, with very little exchange between these two main groups. We have personally observed how growers have transferred cape gooseberry seeds between neighboring departments to establish new cultivated areas in order to overcome biotic and abiotic pressures found during the production cycle (personal observations in Cundinamarca and Boyaca). The results also show that, although there is some degree of differentiation among the cultivated and non-cultivated accessions of the same department, this level of differentiation is not as important as the one observed among the departments or cordilleras. This may suggest that cape gooseberry cultivation activities may be initiated either from cultivated or non-cultivated materials originating in the same or in neighboring departments.
These results of the genetic structure, mainly by geographic region, contrast with the study by Bonilla et al. (2008), who investigated the genetic structure of a collection of the Colombian cape gooseberry from the southwestern part of the country (departments of Nariño, Cauca and Valle del Cauca), Caldas and Cundinamarca. The authors did not observe any relationship between the genetic structure and geography, maybe because their sampling was mainly focused on the departments of the southwest of Colombia (western Andes, group 2 in this study), with very few ac- cessions from the central and eastern Andes. The present results also contrast with the studies by Osorio-Guarínet al. (2016) and Garzón-Martínezet al. (2015) who did not find any relationship between the genetic structure and geography but rather with the biological status. Osorio- Guarínet al. (2016) found three groups of accessions, one group containing mainly wild accessions, another group with cultivated accessions from traditional farmers, and a third group with accessions from commercial cultivated fields. One important observation was that, in the study by Osorio-Guarínet al. (2016), most of the wild genotypes came from the departments of Cundinamarca, Boyaca, Santander and Norte de Santander (eastern Andes), most of the materials from traditional farmers came from Antioquia (central Andes), and most of the commercial varieties came from Cundinamarca and Boyaca (eastern Andes), which could explain why the clustering pattern was not clearly related to geography. In the study by Garzón- Martínezet al. (2015), the number of analyzed accessions was half of those analyzed here, with a poorer geographical representation compared with the present study.
One interesting result of the present study was the large average genetic distance of the non-cultivated accession from Cauca, as can be seen in Fig. 2. However, the fact that Cauca was represented by only one accession did not allow us to reach any conclusion.
These results are certainly important for future breeding activities for the cape gooseberry. It is recommended that future evaluations of germplasm for agronomical traits and disease resistant traits should include wild and cultivated accessions from the two groups identified in this study because some traits might be present in one of the groups and absent in the other. For example, Osorio-Guarínet al. (2016) assessed resistance to F. oxysporum in a set of 100 cape gooseberry cultivated and non-cultivated accessions from different regions in Colombia and found 16 accessions that showed a tolerance response to the fungus, mainly wild accessions from the departments of Antioquia, Cundinamarca and Boyaca.
Conclusions
In general, we can say that the genetic diversity estimators reported here for the Colombian cape gooseberry were lower than those reported in previous studies on the same or related Physalis species. We believe these discrepancies were mainly due to the different genomic coverages in the studies, the type of markers used and the analyzed populations. We can conclude that the Andean cordilleras played a more important role in structuring the genetic diversity in the Colombian cape gooseberry than the biological status (wild versus cultivated). These results have important implications for future studies that aim to evaluate the genetic diversity available in this species and should include samples from the eastern, central and western Andes. The fact that different studies reached different conclusions in relation to genetic structure patterns in the Colombian cape gooseberry leads us to propose that we still need a more comprehensive study with better genomic and geographical sampling.
Acknowledgments
We would like to thank Tulio Cesar Lagos for providing the germplasm material from Nariño that was used in this study. We would also like to thank the funding support received from Ministerio de Agricultura y Desarrollo Rural (Colombia), Contract No. 054/08072-2008L4787-3281.
Literature cited
Agronet. 2015. Cifras agropecuarias. In: www.agronet.gov.co; consulted: December, 2015. [ Links ]
Applied Biosystems. 2006. User bulletin GeneMapper® software version 4.0. Paisley, UK. [ Links ]
Berdugo C., J.A., F. Enciso R., C. González A., and L.S. Barrero M. 2015. Variabilidad genética de parentales y poblaciones F1 inter e intraespecíficas de Physalis peruviana L. y P. f loridana Rydb. Rev. Bras. Frutic. 37, 179-192. Doi: 10.1590/0100-2945-002/14 [ Links ]
Bonilla B., M.L. and K. Espinosa P. 2003. Colección, caracterización fenotípica y molecular de poblaciones de uchuva Physalis peruviana L. Undergraduate thesis. Universidad Nacional de Colombia, Palmira, Colombia. [ Links ]
Bonilla B., M.L., K. Espinosa P., A.M. Posso T., H.D. Vásquez A., and J.E. Muñoz F. 2008. Establecimiento de una colección de trabajo de uchuva del suroccidente colombiano. Acta Agron. 57, 95-99. [ Links ]
Bushakra, J.M., K.S. Lewers, M.E. Staton, T. Zhebentyayeva, and C.A. Saski. 2015. Developing expressed sequence tag libraries and the discovery of simple sequence repeat markers for two species of raspberry (Rubus L.). BMC Plant Biol. 15, 258. Doi: 10.1186/s12870-015-0629-8 [ Links ]
Cailes, R.L. 1952. The cultivation of cape gooseberry. J. Agric. West. Aust. 1, 363-365. [ Links ]
Cavalli-Sforza, L.L. and A.W.F. Edwards. 1967. Phylogenetic analysis: models and estimation procedures. Amer. J. Hum. Genet. 21, 550-570. Doi: 10.2307/2406616 [ Links ]
Council, N.R. 1989. Lost crops of the Incas: little-known plants of the Andes with promise for worldwide cultivation. National Academy Press, Washington D.C. [ Links ]
Criollo E., H., T.C. Lagos B., C.P. Criollo V., and M. Guerrero B. 2001a. Caracterización de materiales de uvilla (Physalis peruviana L.) por sus características de calidad. Rev. Cienc. Agr. 18, 168-180. [ Links ]
Criollo E., H., T. Lagos B., H. Ruiz E., and C. Mosquera Q. 2001b. Evaluación de cultivares de uvilla (Physalis peruviana) con base en su capacidad productiva. Rev. Cienc. Agric. 18, 70-85. [ Links ]
Earl, D.A. and B.M. VonHoldt. 2012. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conservation Genet. Resour. 4, 359-361. Doi: 10.1007/s12686-011-9548-7 [ Links ]
Enciso-Rodríguez, F.E., C. González, E.A. Rodríguez, C.E. López, D. Landsman, L.S. Barrero, and L. Mariño-Ramírez. 2013. Identification of immunity related genes to study the Physalis peruviana - Fusarium oxysporum pathosystem. PloS ONE, 8, e68500. Doi: 10.1371/journal.pone.0068500 [ Links ]
Evanno, G., S. Regnaut, and J. Goudet. 2005. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 14, 2611-2620. Doi:10.1111/j.1365-294X.2005.02553.x [ Links ]
Excoffier, L., P.E. Smouse, and J.M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479-491. [ Links ]
Felsenstein, J. 1993. PHYLIP: phylogenetic inference package, version 3.5 c. University of Washington, Washington DC. [ Links ]
Fischer, G. 2000. Crecimiento y desarrollo. pp. 9-26. In: Flórez, V.J., G. Fischer, and A.D. Sora (eds.). Producción, poscosecha y exportación de la uchuva (Physalis peruviana L.). Unibiblos, Universidad Nacional de Colombia, Bogota. [ Links ]
Fischer, G. and P.J. Almanza-Merchán. 1993. Nuevas tecnologías en el cultivo de la uchuva Physalis peruviana L. Agro-Desarrollo
4, 292-304. [ Links ]
Garzón-Martínez, G.A., Z.I. Zhu, D. Landsman, L.S. Barrero and L. Mariño-Ramírez. 2012. The Physalis peruviana leaf transcriptome: assembly, annotation and gene model prediction. BMC Genom. 13, 151. Doi: 10.1186/1471-2164-13-151 [ Links ]
Garzón-Martínez, G.A., J.A. Osorio-Guarín, P. Delgadillo-Durán, F. Mayorga, F.E. Enciso-Rodríguez, D. Landsman, L. Mariño-Ramírez, and L.S. Barrero. 2015. Genetic diversity and population structure in Physalis peruviana and related taxa based on InDels and SNPs derived from COSII and IRG markers. Plant Gene 4, 29-37. Doi: 10.1016/j.plgene.2015.09.003 [ Links ]
González G., C. and L.S. Barrero M. 2011. Estudio de la marchitez vascular de la uchuva para el mejoramiento genético del cultivo. Corpoica, Mosquera, Colombia. [ Links ]
Herrera M., A.M., J.D. Ortiz A., G. Fischer, and M.I. Chacón S. 2011. Behavior in yield and quality of 54 cape gooseberry (Physalis peruviana L.) accessions from north-eastern Colombia. Agron. Colomb. 29, 189-196. [ Links ]
Herrera M., A.M., G. Fischer, and M.I. Chacón S. 2012. Agronomical evaluation of cape gooseberries (Physalis peruviana L.) from central and north-eastern Colombia. Agron. Colomb. 30, 15-24. [ Links ]
Jakobsson, M. and N.A. Rosenberg. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801-1806. Doi: 10.1093/bioinformatics/btm233 [ Links ]
Juyó, D., F. Sarmiento, M. Álvarez, H. Brochero, C. Gebhardt, and T. Mosquera. 2015. Genetic diversity and population structure in diploid potatoes of group Phureja. Crop Sci. 55, 760-769. Doi: 10.2135/cropsci2014.07.0524 [ Links ]
Kalia, R.K., M.K. Rai, S. Kalia, R. Singh, and A.K. Dhawan. 2011. Microsatellite markers: an overview of the recent progress in plants. Euphytica 177, 309-334. Doi: 10.1007/s10681-010-0286-9 [ Links ]
Lagos B., T.C., H. Criollo E., and C. Mosquera Q. 2001. Evaluación preliminar de cultivares de uvilla (Physalis peruviana L.) para escoger materiales con base en la calidad del fruto. Rev. Cienc. Agric. 18, 82-94. [ Links ]
Lagos B., T.C., F.A. Vallejo C., H. Criollo E., and J.E. Muñoz F. 2008. Biología reproductiva de la uchuva. Acta Agron. 57, 81-87. [ Links ]
Legge, A.P. 1974. Notes on the history, cultivation and uses of Physalis peruviana L. J. Roy. Hort. Soc. 99, 310-314. [ Links ]
Lipka, A.E., F. Tian, Q. Wang, J. Peiffer, M. Li, P.J. Bradbury, M.A. Gore, E.S. Buckler, and Z. Zhang. 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397-2399. Doi: 10.1093/bioinformatics/bts444 [ Links ]
Menzel, M.Y. 1951. The cytotaxonomy and genetics of Physalis. Proc. Amer. Phil. Soc. 95, 132-183. [ Links ]
Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY. [ Links ]
Ortiz, D.C., A. Bohórquez, M.C. Duque, J. Tohme, D. Cuéllar, and T. Mosquera V. 2012. Evaluating purple passion fruit (Passiflora edulis Sims f. edulis) genetic variability in individuals from commercial plantations in Colombia. Genet. Resour. Crop Evol. 59, 1089-1099. Doi: 10.1007/s10722-011-9745-y [ Links ]
Osorio-Guarín, J.A., F.E. Enciso-Rodríguez, C. González, N. Fernández-Pozo, L.A. Mueller, and L.S. Barrero. 2016. Association analysis for disease resistance to Fusarium oxysporum in cape gooseberry (Physalis peruviana L.). BMC Genomics. 17, 248. Doi: 10.1186/s12864-016-2568-7 [ Links ]
Peakall, R. and P.E. Smouse. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and researchan update. Bioinformatics 28, 2537-2539. Doi: 10.1093/bioinformatics/bts460 [ Links ]
Pritchard, J.K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945-959. [ Links ]
Rambaut, A. and A.J. Drummond. 2014. FigTree version 1.4.2. In: http://tree.bio.ed.ac.uk/software/figtree/; consulted: January, 2016. [ Links ]
Rodríguez C., N.C. and M.L. Bueno A. 2006. Study of the cytogenetic diversity of Physalis peruviana L.(Solanaceae). Acta Biol. Colomb. 11, 75-85. [ Links ]
Rosenberg, N.A. 2007. Distruct: a program for the graphical display of structure results. In: https://rosenberglab.stanford.edu/distruct.html; consulted: January, 2016. [ Links ]
Simbaqueba, J., P. Sánchez, E. Sánchez, V.M. Núñez Z., M.I. Chacón, L.S. Barrero, and L. Mariño-Ramírez. 2011. Development and characterization of microsatellite markers for the cape gooseberry Physalis peruviana. PloS ONE 6, e26719. Doi: 10.1371/journal.pone.0026719 [ Links ]
Team, R.C. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [ Links ]
Valencia R., R.A., M. Lobo A., and G.A. Ligarreto M. 2010. Estado del arte de los recursos genéticos vegetales en Colombia: Sistema de Bancos de Germoplasma. Corpoica Cienc. Tecnol. Agropecu. 11, 85-94. [ Links ]
Vega-Vela, N.E. and M.I. Chacón-Sánchez. 2011. Isolation of highquality DNA in 16 aromatic and medicinal Colombian species using silica-based extraction columns. Agron. Colomb. 29, 349-357. [ Links ]
Wei, J., X. Hu, J. Yang, and W. Yang. 2012. Identification of singlecopy orthologous genes between Physalis and Solanum lycopersicum and analysis of genetic diversity in Physalis using molecular markers. PloS ONE 7, e50164. Doi: 10.1371/journal.pone.0050164 [ Links ]














