Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Agronomía Colombiana
versão impressa ISSN 0120-9965
Agron. colomb. vol.34 no.1 Bogotá jan./abr. 2016
https://doi.org/10.15446/agron.colomb.v34n1.51077
Doi: 10.15446/agron.colomb.v34n1.51077
Identification and distribution of whiteflies (Hemiptera: Aleyrodidae) in tomato crops (Solanum lycopersicum) in Cundinamarca (Colombia)
Identificación y distribución de moscas blancas (Hemiptera: Aleyrodidae) en cultivos de tomate de mesa (Solanum lycopersicum) en Cundinamarca (Colombia)
Jorge E. Ángel D.1, Julián Martínez H.1, Maikol Santamaria G.1, Sandra Parada P.1, and Everth Ebratt R.1
1National Phytosanitary Laboratory Diagnosis, Tibaitata Research Center, Instituto Colombiano Agropecuario (ICA). Mosquera (Colombia).jorge.angel@ica.gov.co; eeebraitr@unal.edu.co
Received for publication: 5 June, 2015. Accepted for publication: 28 March, 2016.
ABSTRACT
The main purpose of this study was to determine the presence, distribution and characterization of whiteflies in thirteen tomato-crop producing municipalities in Cundinamarca (Colombia). Immature stages were collected and taken to the laboratory until adults emerged in order to establish their taxonomic identification. The mitochondrial regions were amplified with specific primers, which allowed for the allocation of biotypes in Bemisia tabaci. Genetic similarity analysis was performed in Trialeurodes vaporariorum using RAPD and phylogenetic analysis of the gene sequences mtCOI. The presence of T. vapo- rariorum was established in 100% of the municipalities visited and B. tabaci biotype B was detected in 32%, coexisting with T. vaporariorum. A wide distribution of T. vaporariorum was determined between 653 and 2,680 m a.s.l. B. tabaci was found between 653 and 1,940 m a.s.l distributed in four municipalities in the Sumapaz, lower Magdalena, and Rio Negro provinces. The RAPD analysis established high genetic similarity between the T. vaporariorum insects. The phylogenetic analysis did not allow for the resolution of structured groups inside the analyzed T. vaporariorum samples.
Key words: Trialeurodes vaporariorum, Bemisia tabaci, biotyping, pest insects, Solanaceae, geographical distribution.
RESUMEN
El presente trabajo tuvo como objetivo determinar la presencia, distribución y caracterización de moscas blancas en trece municipios productores del departamento de Cundinamarca en cultivos de tomate de mesa. Se recolectaron estados inmaduros, los cuales fueron llevados al laboratorio hasta la obtención de adultos para la identificación taxonómica de los mismos. La asignación de biotipos en Bemisia tabaci se realizó mediante la amplificación de regiones mitocondriales con iniciadores específicos; se realizó un análisis de similitud genética en Trialeurodes vaporariorum mediante RAPDs y un análisis filogenético de secuencias del gen mtCOI. Se estableció la presencia de T. vaporariorum en el 100% de los municipios visitados y B. tabaci biotipo B en el 32%, coexistiendo con T. vaporariorum. Se determinó una amplia distribución de T. vaporariorum entre los 653 y 2.680 msnm; B. tabaci se encontró entre los 653 y 1.979 msnm, distribuida en cuatro municipios ubicados en las provincias de Sumapaz, bajo Magdalena y Rionegro. Mediante análisis RAPDs se estableció una alta similitud genética entre individuos de T. vaporariorum. El análisis filogenético no permitió la resolución de grupos estructurados dentro de las muestras de T. vaporariorum analizadas.
Palabras clave: Trialeurodes vaporariorum, Bemisia tabaci, biotificación, insectos plaga, Solanaceae, distribución geográfica.
Introduction
The tomato (Solanum lycopersicum L.) is one of the most important crops in Colombia. It is estimated that more than 17,000 families are directly linked to its cultivation (Jara- millo et al., 2006). Trialeurodes vaporariorum (Westwood) and Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) affect the production of this fruit. These whitefly species are considered the most important insect plague in Colombia (Cardona et al., 2005). These insects are efficient vectors in transmitting Crinivirus and Begomovirus and globally limit tomato production (Wintermantel, 2004). Due to the existence of morphologically indistinguishable biotypes in B. tabaci, it is necessary to utilize molecular techniques to identify these biotypes (Mendoza et al., 1995; Polston and Anderson, 1999). Until now, the existence of biotypes has not been reported for T. vaporariorum; similar research has shown no phylogenetic differentiation in T. vaporariorum populations (Roopa, 2012).
Whitefly populations constantly occupy new environments. T. vaporariorum was recorded beginning at 600 m a.s.l. (Rodríguez and Cardona, 2001) up to 3,000 m a.s.l. in the high tropics in the Andean region (Rendónet al., 2001). The biotype B from B. tabaci has been recorded between 383 and 1,857 m a.s.l., sharing an ecological niche with T. vaporariorum from 832 m a.s.l. (Martínezet al., 2012).
Due to the importance of the whitefly as a viral vector, the large number of affected agriculturally important host plant species and the economic losses in vegetable and flower crops (Agrios, 2005), along with its wide altitudinal and potential distribution in Cundinamarca, it is necessary to conduct further whitefly species inspection and detection studies in the principal tomato production areas of Colombia. This study aimed to determine the current status of T. vaporariorum and B. tabaci in Cundinamarca by identifying its biotypes and analyzing the genetic polymorphisms obtained through RAPDs and mtCOI sequences in T. vaporariorum.
Materials and methods
Sampling zones. Samples were collected from tomato crops in greenhouses and open-air exposed areas, in the vegetative or reproductive stages, in thirteen municipalities in Cundinamarca located between 653 and 2,680 m a.s.l. Coordinates were obtained from each farm (GPS 40 Garmin®, Lenexa, KS) as well as information on the host range of tomato crop growth stage and crop type (Tab. 1). The farm selection for sampling the tomato crops was per- formed according to information provided by the Instituto Colombiano Agropecuario (ICA) and regional farmers in each municipality. The number of sampling farms varied in some municipalities depending on the availability of tomato crops for analysis at the time of sampling.
Severity of whitefly infestation. Approximately 1,000 m2 of each tomato crop was inspected and 50 plants were selected at random. The infestation was determined according to the number of plants with whitefly nymphs or adults in relation to the number of plants observed.
% Infestation = total number of affected plants/total number of plants (Eq. 1)
To determine the severity, the middle third leaf was chosen on each plant and the approximate area occupied by the underside of the whitefly nymphs was established (Cardona et al., 2005).
% Severity = number four nymphal stage per leaflet/ total number of nymphs observed per leaflet (Eq. 2)
Retrieval of adult whiteflies. Seven plants were chosen from each farm in which two leaves with whitefly nymphs were collected. The samples were labeled and transported in styrofoam boxes at 5°C until being stored at 4°C.
The taxonomic identity of the whiteflies was established by analyzing the fourth instar nymph (Caballero, 1994). The leaves of the nymphs were individually prepared on Petri dishes with a substrate of absorbent paper moistened with distilled water. The emerged whitefly adults were fed a 2% sugar solution (w/v). Afterward, they were sexed and stored in 1.5 mL Ependorff tubes in 95% ethanol at -20°C.
Extraction of DNA from whiteflies. Each female individual was soaked in 40 μL of lysis buffer (5 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 0.5% Nonidet P-40, 1 mg mL-1proteinase K) in PCR tubes. The lysate was incubated for 15 min at 65°C and 10 min at 95°C. The supernatant was stored at -20°C until its use (Frohlich et al., 1999).
Identification of Bemisia tabaci biotypes. Primers for specific detection of biotype B and Q were used (Shatters et al., 2009). The mixing conditions and amplification were performed in accordance with those reported by Shatters et al. (2009) in a final volume of 12.5 uL of 1 U of Taq polymerase. The PCR reaction was completed by a thermocycler PT200 (MJ Research, Watertown, MA) using reagents from Invitrogen. The results were corroborated by RAPDs with an OPA4 primer (Martínezet al., 2012).
Genetic variability analysis of T. vaporariorum by RAPDs. The DNA amplification of 25 T. vaporariorum individual females was carried out with RAPDs primers: OPA4, OPA9, and OPC2 (Salas and Arnal, 2001; Martínezet al., 2012). The reproducibility of the technique was determined by parameters proposed by Pérez et al. (1998) on three samples amplified five times.
The genetic similarity was determined by the Dice similarity coefficient (Sneath et al., 1975) and a dendrogram was constructed using the UPGMA algorithm. The genetic similarity matrix was correlated with the matrix of geo- graphical distances of the samples analyzed by the Mantel test (Mantel, 1967). The analysis was performed with the PAST program, version 1.34 (Hammer et al., 2001).
T. vaporariorum phylogenetic analysis. A 49 sequences were selected for the phylogenetic reconstruction. A 35 accessions of T. vaporariorum, T. ricini, and T. lauri were downloaded from the NCBI (www.ncbi.nlm.nih.gov/genbank/) and 14 came from the amplification and sequencing of mitochondrial products obtained with primers C1-J-2195 and TL2-N-3014 (Simon et al., 1994). The obtained sequences were indexed in the NCBI database. Sequence alignment was performed with MUSCLE and the phylogenetic analysis was performed with MEGA.5.2 (Tamura et al., 2011). A phylogenetic tree with the available sequences was constructed using the maximum parsimony method, excluding gaps analysis with 1,000 boopstrap replicates, using the nucleotide substitution model and the Subtree-Pruning-Regrafting (SPR) search method for tree inference.
Based on the phylogenetic results, the generated clades were selected to estimate the distances of evolutionary divergence between the group means; the analysis was conducted using the Kimura-2-parameter (K2P) algorithm as a model of distance (Kimura, 1980; Saitou and Nei, 1987), eliminating all positions with gaps (Chu et al., 2010) with 1,000 bootstraps using MEGA.5.2 (Tamura et al., 2011).
Discussion and results
Whitefly distribution. T. vaporariorum was found in 100% of the assessed farms distributed between 653 and 2680 m a.s.l. in tomato crops in greenhouses and open-air exposed areas in the Tibacuy and Caqueza municipalities, respectively. Berrío (2007) and Martínezet al. (2012) recorded T. vaporariorum in the La Vega, San Francisco, Choachi, Fomeque, Ubaque, and Caqueza municipalities.
B. tabaci was recorded in coexistence with T. vaporariorum in 32% of the farms tested in the Fusagasuga, Guaduas, Pacho, and Tibacuy municipalities. It was distributed at altitudes between 653 and 1,940 m a.s.l. in greenhouse and open-air exposed crops (Tab. 1). Martínezet al. (2012) recorded the coexistence of both species up to an altitude of 1,857 m a.s.l. in several municipalities in Cundinamarca. In Colombia, the coexistence T. vaporariorum and B. tabaci was observed in the Tolima, Huila, and Valle del Cauca departments (Quintero et al., 2001).
Infestation and severity. The highest percentages of infestation occurred when T. vaporariorum was recorded as a single species. In the municipalities of Cundinamarca, where a great number of tomato farms reside, the aver- age percentages of infestation were recorded between 46 to 100% and severity between 8.8 and 80%, respectively. In contrast, the average percentages of infestation were recorded from 40.25 to 69.80% and the severity from 25 to 49%, respectively, in the properties located in municipalities where T. vaporariorum and B. tabaci coexisted (Tab. 2). Frequent assessments are required to determine the influence of the coexistence of T. vaporariorum and B. tabaci in rates of infestation and severity.
Bemisia tabaci biotypes. In all of the individuals it was possible to unambiguously assign the identity of biotypes. Amplicons of expected sizes 478 and 303 pb were obtained respectively for biotype B and Q (Fig. 1). A total of 52 samples of B. tabaci from the Fusagasuga, Guaduas, Pacho, and Tibacuy municipalities were analyzed to identify biotypes. All samples corresponded to B. tabaci biotype B.

In a study conducted in the Valle del Cauca department, the dominance of B. tabaci biotype B was observed in different crops (Rodríguez et al., 2005). Similarly, in 2012, no evidence of the presence of biotype A was found in 12 municipalities in the same department (Rodríguez et al., 2012). In Cundinamarca, a recent study (Martínezet al., 2012) reinforced the findings in this research where the dominance of B. tabaci biotype B was determined (Rodríguez et al., 2012). In the analyzed samples, the presence of biotype Q was not detected.
Genetic variability of T. vaporariorum. The RAPD-PCR patterns with OPA2, OPC2, and OPA4 primers were consistent in comparing the electrophoretic profiles between repetitions. The parameters for assessing the reproducibility of this technique were very similar with all three primers (Tab. 3), which indicates the feasibility of their use for polymorphism analysis (Pérez et al., 1998).

In Fig. 2, the electrophoretic patterns of the amplifications using the OPC-2 primer are shown. Similar amplification profiles are evident in samples tested as amplifications with OPA-9 and OPA-4 primers (figures not shown).
As shown in the dendrogram (Fig. 3), the samples are grouped with a percentage of similarity greater than 70%, which indicates a high genetic similarity among the analyzed individuals. The formation of two groups is evidenced by more than 80% similarity, comprised of samples from different municipalities. The groups do not reflect the origin of the samples (Tab. 4).
By calculating the linear correlation between Dice similarity matrices and geographical distances, a slight correlation r (A, B) of 0.2 (p-value <0.005; alpha = 0.05) was determined, which indicates a slight trend of an increase in genetic distances between individuals analyzed with respect to geographical distances.
Phylogenetic analysis of T. vaporariorum. Diversity studies in agriculturally important insects have been used to identify new species, biotypes, and haplotypes, which are difficult to identify by morphological characteristics (Perring, 2001; Ball and Armstrong, 2006). The Cytochrome Oxidase I gene (mtCOI) has been widely used as a molecular marker to identify whitefly species and its variants that exhibit biological differences but no morphological differences (Frohlich et al., 1999; Maruthi et al., 2007). In this study, the topology of the phylogenetic analysis (Fig. 4) shows the formation of clades, separat- ing T. vaporariorum accessions of T. ricini and T. lauri species, which have been reported as phylogenetically similar (Roopa et al., 2012).
The phylogenetic tree was divided into three highly supported clades by bootstrap separating the accessions of T. vaporariorum comprised of groups of samples from America and Asia (clade I) with a support of 100%, T. lauri (clade II), and T. ricini (clade III). Within clade I, accessions from Costa Rica and Colombia are presented in the American group and the Asian samples from India and China formed the T. vaporariorum clade. Although the samples from America and Asia appear in distinct clusters, the topology support by bootstrap analysis was 60%. T. ricini and T. lauri appeared in this analysis as sister groups (100% bootstrap) and samples of each species also appeared as monophyletic groups and supports of 100%.
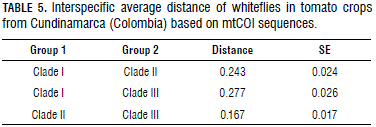
The estimation of evolutionary divergence distances between the groups (Tab. 5) showed a distance of 24% between clade I that grouped the samples of T. vaporariorum and clade II with the samples T. lauri. The difference of the evolutionary distance between the T. lauri and T. ricini species was 16%, which confirms the findings reported by Chu et al. (2010), where high phylogenetic closeness between the two species was determined.
Studies of genetic diversity and phylogenetic reconstructions are virtually nonexistent in T. vaporariorum, as compared with B. tabaci, reported as a species complex in which 24 biotypes have been proposed to exist (De Barro et al., 2011) although more detailed studies of reproductive isolation reduce this number to less than half.
In the study conducted by Roopa et al. (2012), in which a phylogenetic analysis of various species of the genus Trialeurodes sp. was performed, no genetic structuring within the T. vaporariorum species was determined. The analysis conducted based on the sequences of mitochondrial and nuclear genes did not allow for the differentiation of any group in the phylogenetic reconstructions, from which it was concluded that this species does not constitute the species complex.
In this study, the sequences of T. vaporariorum were from the American and Asian continents. Clearly, a phylogenetic separation between the American samples from Colombia and Costa Rica and from the Asian continent is provided although the node support between the groups was low, which may be due to the resolution of the gene or sample size. In this research, the existence of genetic structuring within T. vaporariorum was not apparent, far from suggesting the presence of biotypes in this species, but it exposes the need for further studies to elucidate the phylogenetic and ecological processes concerning this species on which so little information has been extracted.
Conclusions
A wide distribution of T. vaporariorum and the presence of B. tabaci were recorded in four municipalities of Cundinamarca. T. vaporariorum and B. tabaci coexisted in the Fusagasuga, Tibacuy, Pacho, and Guaduas municipalities. The infestation and severity in the tomato was higher when T. vaporariorum was presented as a single species. This finding is in accordance with previous observations that report whitefly species in overlapping niches, which have demonstrated a displacement of T. vaporariorum by B. tabaci in interspecific competitive interactions (Zhang et al., 2011). The increase of the biotic potential of whiteflies, depending on the interactions, is important in terms of virus epidemiology due to its role as a vector of plant vi- ruses (Wintermantel and Hladky, 2010; Navas-Castillo et al., 2011). Specimens collected from different geographical regions of Cundinamarca showed high genetic similarity, >75%. In this study, the presence of possible biotypes was not evidenced in T. vaporariorum and, although a differentiation of the two groups within the T. vaporariorum clade was observed, it did not allow for assertions to be made about the genetic structure of the species at this level.
Acknowledgements
This publication is a product of research project code: 2106- 521-28363, financed by Colciencias, Contract Number: CTB796-2011, executed by the Instituto Colombiano Agropecuario - ICA through the Deputy Manager of Analysis and Diagnosis, Management of Technical Agriculture, National Plant Health Diagnostic Laboratory CI Tibaitata.
Literature cited
Agrios, G.N. 2005. Plant pathology. 5th ed. Academic Press, Elsevier, Burlington, MA. [ Links ]
Ball, S.L. and K.F. Armstrong. 2006. DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae). Can. J. For. Res. 36, 337-350. Doi: 10.1139/x05-276 [ Links ]
Berrío R., M.P. 2007. Identificación y distribución de especies y biotipos de moscas blancas sobre hortalizas en Cundinamarca. Undergraduate thesis. Faculty of Sciences, Pontificia Universidad Javeriana, Bogota. [ Links ]
Caballero, R. 1994. Clave de campo para inmaduros de mosca blancas en Centroamérica (Homóptera: Aleyrodidae). Ceiba 35, 47-51. [ Links ]
Cardona, C., A. López-Ávila, and O. Valarezco. 2005. Colombia and Ecuador. pp. 274-284. In: Anderson, P.K. and F.J. Morales (eds.). Whitefly and whitefly-borne viruses in the tropics: building a knowledge base for global action. Centro Internacional de Agricultura Tropical (CIAT), Cali, Colombia. [ Links ]
Chu, D., G. Liu, F. Wan, Y. Tao, and R.J. Gill. 2010. Phylogenetic analysis and rapid identification of the whitefly, Bemisia afer, in China. J. Insect Sci. 10, 86.Doi: 10.1673/031.010.8601 [ Links ]
De Barro, P.J., S.S. Liu, L.M. Boykin, and A.B. Dinsdale. 2011. Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 56, 1-19. Doi: 10.1146/annurev-ento-112408-085504 [ Links ]
Frohlich, D.R., I. Torres-Jerez, I.D. Bedford, P.G. Markham, and J.K. Brown. 1999. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 8, 1683-1691. Doi: 10.1046/j.1365-294x.1999.00754.x [ Links ]
Hammer, Ø., D.A.T. Harper, and P.D. Ryan. 2001. PAST: paleontological statistics software package for education and data analysis. Paleontol. Electron. 4, 9. [ Links ]
Jaramillo N., J., V.P. Rodríguez, M. Guzmán A., and M.A. Zapata. 2006. El cultivo del tomate bajo invernadero (Lycopersicon esculentum Mill.). Boletin Tecnico No. 21. Corpoica, Rionegro, Colombia. [ Links ]
Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol.16, 111-120. Doi: 10.1007/BF01731581 [ Links ]
Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res 27, 209-220. [ Links ]
Martínez B., O.Y., E.E. Ebratt R., W. Turizo A., O. Guerrero G., and R. Acosta A. 2012. Presence of Bemisia tabaci (Hemiptera: Aleyrodidae) and Begomovirus, associated with tomato crops Solanum lycopersicum L. in Cundinamarca. Agron. Colomb. 30, 395-402. [ Links ]
Maruthi, M.N., A.R. Rekha, P. Sseruwagi, and R.J. Hillocks. 2007. Mitochondrial DNA variability and development of a PCR diagnostic test for populations of the whitefly Bemisia afer (priesner and hosny). Mol. Biotechnol. 35, 31-40. Doi: 10.1385/MB:35:1:31 [ Links ]
Mendoza, J., M. Arias, R. Quijije, E. Cañarte, and V. Álvarez. 1995. Reporte de Ecuador. Memorias IV taller latinoamericano sobre moscas blancas y geminivirus. Ceiba 36, 13-15. [ Links ]
Navas-Castillo, J., E. Fiallo-Olivé, and S. Sánchez-Campos. 2011. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219-248. Doi: 10.1146/annurev-phyto-072910-095235 [ Links ]
Pérez, T., J. Albornoz, and A. Domínguez. 1998. An evaluation of RAPD fragment reproducibility and nature. Mol. Ecol. 7, 1347-1357. Doi: 10.1046/j.1365-294x.1998.00484.x [ Links ]
Perring, T.M. 2001. The Bemisia tabaci species complex. Crop Prot. 20, 725-737. Doi: 10.1016/S0261-2194(01)00109-0 [ Links ]
Polston, J. and P. Anderson. 1999. Surgimiento y distribución de geminivirus transmitidos por mosca blanca en tomate en el Hemisferio Occidental. Manejo Integrado de Plagas 53, 24-42. [ Links ]
Quintero, C., F. Rendón, J. García, C. Cardona, A. López-Ávila, and P. Hernández. 2001. Especies y biotipos de moscas blancas (Homoptera: Aleyrodidae) en cultivos semestrales de Colombia y Ecuador. Rev. Colomb. Entomol. 27, 27-31. [ Links ]
Rendón, F., C. Cardona, and J.M. Bueno. 2001. Pérdidas causadas por Trialeurodes vaporariorum (Homoptera: Aleyrodidae) y Thrips palmi (Thysanoptera: Thripidae) en habichuela en el Valle de Cauca. Rev. Colomb. Entomol. 27, 39-43. [ Links ]
Rodríguez, I.V. and C. Cardona. 2001. Problemática de Trialeurodes vaporariorum y Bemisia tabaci (Homoptera: Aleyrodidae) como plagas de cultivos semestrales en el Valle del Cauca. Rev. Colomb. Entomol. 27, 21-26. [ Links ]
Rodríguez, I., H. Morales, J.M. Bueno and C. Cardona M. 2005. El biotipo B de Bemisia tabaco (Homoptera: Aleyrodidae) adquiere mayor importancia en el Valle del Cauca. Rev. Colomb. Entomol. 31, 21-28. [ Links ]
Rodríguez T., I., J.M. Bueno M., C. Cardona M., and H. Morales M. 2012. Biotipo B de Bemisia tabaco (Hemiptera: Aleyrodidae): plaga de pimentón en el Valle del Cauca, Colombia. Rev. Colomb. Entomol. 38, 14-22. [ Links ]
Roopa, H.K., N.K. Krishna Kumar, R. Asokan, K.B. Rebijith, R. Mahmood, and A. Verghese. 2012. Phylogenetic analysis of Trialeurodes spp. (Hemiptera: Aleyrodidae) from India based on differences in mitochondrial and nuclear DNA. Fla. Entomol. 95, 1086-1094. Doi: 10.1653/024.095.0438 [ Links ]
Saitou, N. and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406-425. [ Links ]
Salas, J. and E. Arnal. 2001. Bemisia tabaco (Gennadius, 1899) biotipo B, primer registro para Venezuela utilizando RAPD's- PCR. Entomotropica 16, 181-185. [ Links ]
Shatters, R.G., C.A. Powell, L.M. Boykin, H. Liansheng, and C.L. Mckenzie. 2009. Improved DNA barcoding method for Bemisia tabaci and related Aleyrodidae: development of universal and Bemisia tabaci biotype-specific mitochondrial cytochrome c oxidase I polymerase chain reaction primers. J. Econ. Entomol. 102, 750-758. Doi: 10.1603/029.102.0236 [ Links ]
Simon, C., F. Frati, A. Beckenbach, B. Crespi, H. Liu, and P. Flook. 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651-701. Doi: 10.1093/aesa/87.6.651 [ Links ]
Sneath, P.H.A., R.R. Sokal and W.H. Freeman. 1975. Numerical taxonomy. The principles and practice of numerical classification. Syst. Zool. 24, 263-268. Doi: 10.2307/2412767 [ Links ]
Tamura, K., D. Peterson, N. Peterson, G. Stecher, M. Nei, and S. Kumar. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731-2739. Doi: 10.1093/molbev/msr121 [ Links ]
Wintermantel, W. 2004. Emergence of greenhouse whitefly (Trialeurodes vaporariorum) transmitted criniviruses as threats to vegetable and fruit production in North America. In: APSnet Feature, www.apsnet.org/publications/apsnetfeatures/Documents/2004/GreenhouseWhitefly.pdf; consulted: March, 2016. [ Links ]
Wintermantel, W.M. and L.L. Hladky. 2010. Methods for detection and differentiation of existing and new crinivirus species through multiplex and degenerate primer RT-PCR. J. Virol. Methods 170, 106-114. Doi: 10.1016/j.jviromet.2010.09.008 [ Links ]
Zhang, G.-F., D.-C. Li, T.-X. Liu, F.-H. Wan, and J.-J. Wang. 2011. Interspecific interactions between Bemisia tabaci biotype B and Trialeurodes vaporariorum (Hemiptera: Aleyrodidae). Environ. Entomol. 40, 140-150. Doi: 10.1603/EN10135 [ Links ]














