Introduction
Endornavirus is a recently approved (2006) genus within the family Endornaviridae that comprises viruses characterized by a naked double-stranded RNA (dsRNA) genome of about 9.8-17.6 kb, a single open reading frame (ORF) and a site-specific single chain break (nick) about 1.2-2.7 kbp from the 5' end (King et al., 2012). The encoded polypeptide is a large multidomain protein of about 3,000-6,000 amino acids that may contain methyltransferase (MT), viral RNA helicase (Hel), glucosyltransferase (GT) and RNA-dependent RNA polymerase (RdRp) domains; only the latter is observed across all species (Roossinck et al., 2011). Endornaviruses are normally found in low copy numbers (~100 copies/cell), seem to lack cell-to-cell movement and, apparently, can only be transmitted vertically through seeds and pollen in plants and spores in fungi or oomycetes (Straminipila) (Roossinck et al., 2011). In general, endornaviruses do not cause visible disease symp-toms (Gibbs et al., 2000; Fukuhara et al., 2006) but some evidence suggests their involvement in cytoplasmic male sterility in broad bean (Viciafaba) (Grill and Garger, 1981; Pfeiffer, 1998) and hypovirulence in the plant pathogenic fungi Helicobasidium mompa (Osaki et al., 2006).
The first plant endornavirus genome sequence was obtained for Oryza sativa endornavirus (OsEV) identified in cultivated rice (Oryza sativa ssp. japonica), which was to become the type species of the genus (Moriyama et al., 1995; Fukuhara and Moriyama, 2008). The existence of endornaviruses infecting other plants was subsequently confirmed in wild rice (Oryza rufipogon; Moriyama et al., 1999), broad bean (Vicia fava;Pfeiffer, 1998), kidney bean (Phaseolus vulgaris;Wakarchuk and Hamilton, 1990) barley (Hordeum vulgare;Zabalgogeazcoa and Gildow, 1992), Yerba mate (Ilexparaguariensis;Debat et al., 2014), Malabar spinach (Basella alba L.; Okada et al., 2014), Grapevine (Vitis vinifera; Espach et al., 2012), Avocado (Persea Americana;Villanueva et al., 2012), cucurbits (Coutts, 2005) and various Capsicum species (Valverde and Gutiérrez, 2007; Jo et al., 2015). As indicated, endornaviruses are not exclusive to plants as they have been found infecting basidiomycetes: Rhizoctonia solani (Das et al., 2014), Rhizoctonia cerealis (Li et al., 2014), Helicobasidium mompa (Osaki et al., 2006); ascomycetes: Rosellinia necatrix (Yaegashi and Kanematsu, 2016), Erysiphe cichoracearum (Du et al., 2016), Alternaria brassicicola (Shang et al., 2015), Sclerotinia sclerotiorum (Khalifa and Pearson, 2014), Tuber aestivum (Stielow et al., 2011), Gremmiella abietina (Tuomivirta et al., 2009) and oomycetes within the genus Phytophthora (Hacker et al., 2005; Kozlakidis et al., 2010).
The first report regarding endornaviruses in Capsicum annuum described them as large molecular weight dsRNAs present in chloroplast fractions (Valverde et al., 1990), however, these molecules were only classified as the genomes of endornaviruses 17 years later after cDNA sequencing revealed their phylogenetic affinity to Oryza sativa endor-navirus (OsEV) and Oryza rufipogon endornavirus (OrEV) in 2007 (Valverde and Gutiérrez, 2007). Endornaviruses are ubiquitous in Capsicum species (Okada et al., 2011), as confirmed by in silico analysis of pepper transcriptomes deposited in public databases (Jo et al., 2016) and Nextgeneration sequencing (NGS) studies of total small RNAs (Sela et al., 2012; Chen et al., 2015; Jo et al., 2015; Lim et al., 2015). In this work, we report the sequence of two Bell pepper endornavirus (BPEV) genomes naturally infecting Capsicum annuum in the province of Antioquia (Colombia). RT-PCR, RT-qPCR and Sanger sequencing using primers designed for this purpose confirmed the presence of BPEV in field samples.
Materials and methods
Next-generation sequencing
High-throughput sequencing of the C. annuum transcrip-tomes was performed on bulk leaf samples of chili pepper collected at the municipalities of Santa Fe de Antioquia and bell pepper from El Peñol (Antioquia, Colombia). After grinding the leaf tissue with liquid nitrogen, total RNA was extracted with the GeneJET Plant RNA Purification Mini kit (Thermo Fisher Scientific, USA) and rRNA depleted with the TruSeq Stranded Total RNA with Ribo-Zero Plant kit (Illumina, USA). Libraries were constructed using the TruSeq RNA Sample Preparation kit (Illumina, USA) and sequencing performed with the Illumina HiSeq 2000 system service provided by Macrogen (South Korea). Adapter sequences and low quality bases were removed with SeqTK prior analysis (https://github.com/lh3/seqtk). Sequence assembly was performed with Trinity (Grabherr et al., 2011) and endornaviral contigs were identified by a local BLASTN search using a database of endornavirus reference genomes. Genome assemblies were confirmed by mapping with Bowtie2 (Langmead and Salzberg, 2012) and checked for inconsistencies and assembly artifacts with Tablet (Milne et al., 2010). Consensus sequences were deposited in GenBank under accession codes KX977568 (BPEV from Bell pepper) and KX977569 (BPEV from Chili pepper).
RT-PCR and RT-qPCR detection
Primers BPEV_F (5'-AGG CTA AAT GTG CAC CTA AAA TTG G-3'; Tm = 60.3°C), BPEV_R (5'-TTT CTC AGC GAC TGC TGA CC-3'; Tm = 60.3°C) and qBPEV_R (5'-CTT TAC ACT GCC ATA ACA ACG C-3'; Tm = 58.5°C) were designed for specific amplification of BPEV using RT-PCR (BPEV_F and BPEV_R) or RT-qPCR (BPEV_F and qBPEV_R) using the assembled genomes as reference. Primer specificity was verified in silico using the program primer-BLAST (Ye et al., 2012). Experimental validation of primers was performed in ten random leaf samples collected in four commercial bell pepper fields from the municipalities of Marinilla and El Peñol (Antioquia). The NGS samples and a reaction mix lacking template cDNA were used as positive and negative controls, respectively. RNA was extracted from 100 mg of ground tissue using the Gene-JET Plant RNA Purification kit (Thermo Fisher Scientific, Waltham, MA, USA) and eluted in 40 µL of DEPC treated water; the purity and concentration were determined by absorbance readings at 260 and 280 nm using a Nanodrop 2000C (Thermo Fisher Scientific). Retrotranscription was performed for 30 min at 50°C in 20 µL containing 200 U of Maxima Reverse Transcriptase (Thermo Fisher Scientific), 1X RT Buffer, 0.5 mM dNTP Mix, 100 pmol of specific reverse primer BPEV_R, 20 U de RiboLock RNase Inhibitor and 100-500 ng of total RNA. For the qPCR, the Maxima SYBR Green/ROX qPCR Master Mix (2X) kit (Thermo Fisher Scientific) was used in 25 µL of reaction containing 12.5 µL mix, 10 µL DEPC water, primers BPEV_F/ qBPEV_R at 0.3 µM and 50-100 ng cDNA. Samples were considered positive if they exhibited fluorescence values higher than the threshold before the 35th cycle (Schena et al., 2004). Primer specificity was verified by High Resolution Melting (HRM) in the 50 and 99°C range and compared to melting temperature (Tm) values from the positive controls (NGS samples). RT-PCR validation of primers BPEV_F/BPEV_R was performed using cDNA obtained as described above. The amplification mix included 17.8 µL water, 1X enzyme buffer, 1.8 mM MgCl2, 0.2 mM dNTPs, 0.2 µM primers, 1 U Taq DNA polymerase (Thermo Fisher Scientific, USA) and 50-100 ng de cDNA in a final volume of 25 µL. The amplification consisted of an initial 3 min incubation at 95°C, followed by 35 cycles that included denaturation at 94°C (30 s), annealing at 52°C (1 min) and extension at 72°C (1 min); the amplification ended with an extension cycle at 72°C for 5 min. Amplicon size was determined by 1.8% agarose gel electrophoresis stained with GelRed 1X (Biotium, Hayward, CA, USA) in Bio Doc Analyze transilluminator (Biometra, Góttingen, Germany). Some sequences for qRT-PCR and RT-PCR amplification products were confirmed by the Sanger method using an ABI Prism 3730xl Sequencer (Applied Biosystems, Carlsbad, CA, USA) at Macrogen (South Korea). Sanger sequences were deposited in GenBank under accession codes KX977563-KX977567.
Bioinformatic analyses
The ORF coding for the endornaviral polyprotein was identified using the DNA translate program available at http://web.expasy.org. Identification of protein motifs was performed in the Conserved Domain Database at NCBI (Marchler-Bauer et al., 2015). Evolutionary analyses based on the complete polyprotein were constructed by the Jones-Taylor-Thornton model using a gamma distribution with five categories and a shape parameter of 1.09 (Jones et al., 1992). Evolutionary distances for the partial nucleotide sequences were computed using the Tamura 3-parameter method, rate variation among sites was modeled with a gamma distribution with a shape parameter of 1.2. Phylogenetic trees were calculated in MEGA7 using the Neighbor-Joining method with 1,000 bootstrap replicates (Kumar et al., 2016).
Results and discussion
Next-generation sequencing of the C. annuum sample of chili pepper from Santa Fe de Antioquia resulted in a transcriptome of 5,605,703 paired-end reads with a total of 1,121,140,600 nt. A local BLASTN search identified 1,033 reads (0.018%) with significant similarity to Bell pepper endornavirus (BPEV, NC_015781) with percent sequence identities between 82.5 and 100 percent (Fig. 1A). Similar results were obtained with the sample of bell pepper from El Peñol. In this case, a transcriptome consisting of 6,901,644 paired-end reads (1,380,328,800 nt) was obtained of which 2,936 were classified as associated to the BPEV genome (0.042%), with sequence identities ranging from 82.2 to 100 per cent (average of 95.2%) (Fig. 1B). Previous studies have shown the copy number of endornaviral sequences in C. annuum samples to be in the 0.01% to 0.18% range, which coincides with our results (Jo et al., 2016). As the species demarcation criteria within the genus Endornavirus has been set below 75% nucleotide sequence identity (King et al., 2012), it is clear that both viruses are BPEV isolates.
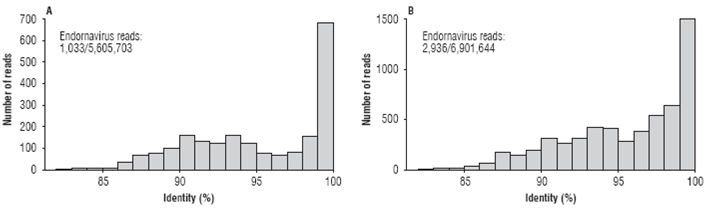
FIGURE 1 Frequency histograms showing the number of paired-end reads at different nucleotide sequence percent identity with respect to member of the genus Endornavirus in the Capsicum annuum transcriptomes from the municipalities of Santa Fe de Antioquia (A) and El Peñol (B) in Antioquia (Colombia).
After confirming the presence of endornavirus in both samples, their genomes were assembled. The BPEV se-quence from Santa Fe de Antioquia (BPEV_Santa_Fe) resulted in a consensus of 14,727 nt with an average sequence depth of 48.25X and 34 polymorphic sites (Fig. 2A). BPEV_Santa_Fe encodes a putative protein of 4,885 amino acids between nucleotide positions 26 to 14,683 that contains viral methyltransferase (M, cl03298), viral RNA helicase (H, pfam01443), glycosyltransferase (GT,cd03784) and RNA-dependent RNA polymerase (RdRp,cl03049) motifs at amino acid positions 327-562 (e-value: 3.98e-8), 1413-1649, (e-value: 2.23x10-6), 3,113-3,458 (e-value: 1.17x10-25) and 447-4,785 (e-value: 1.14x10-21), respectively. Amino acid polymorphisms were observed at positions S293G, E813D, V823F, N1118D, E1129D, S1214L, E1720D, K1776R, I1887L, K3919R and S4350.
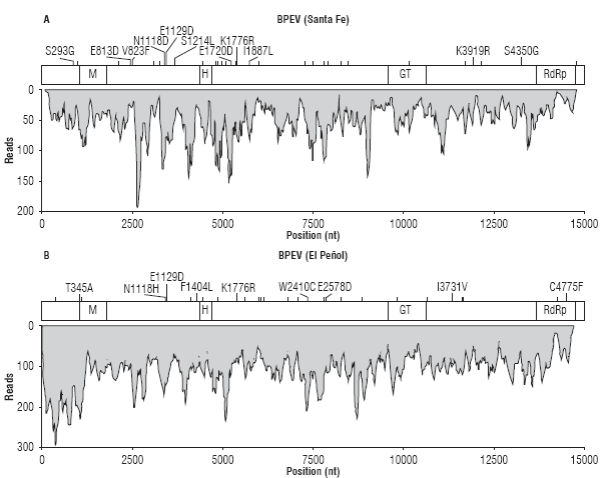
FIGURE 2 Sequence coverage and variability of the BPEV assemblies from Santa Fe de Antioquia (A) and El Peñol (B). The position of the open reading frame encoding for the endornaviral polyprotein is shown on top of each coverage graph and includes the location of predicted methyltransferase (M), helicase (H), glucosyltransferase (GT) and RNA-dependent RNA polymerase (RdRp) domains. Nucleotide polymorphic sites are indicated as vertical bars; amino acid changes are explicitly indicated when applicable.
The sample from El Peñol resulted in a contig of 14,714 nt (BPEV_Peñol) with average sequence depth of 105.4X (max 290X) and 32 polymorphic positions (Fig. 2B). An ORF, encoding a protein of 4,884 residues with a similar domain structure as BPEV_Santa_Fe was identified between positions 20 and 14,674. The following amino acid sequence variations resulting from polymorphisms in the assembly were observed: T345A, N1118H, E1129D, F1404L, K1776R, W2410C, E2578D, I3731V, C4775F. The consensus genomes of BPEV_Santa_Fe and BPEV_Peñol were practically identical as only eight nucleotide changes (T23C, T3647C, C3666T, T4826C, A5684T, A7720G,C10033T and G11781A) and three amino acid substitutions (L1214S, L1887I, K3919R) were observed; additionally, a leucine codon (GCT) is inserted in BPEV_Santa_Fe at position 11,464.
A survey of GenBank sequences using BPEV_Santa_Fe and BPEV_Peñol as query, resulted in significant matches with BPEV isolates Kyosuzu from Japan (99.2%, AB597230), Maor (99.3%, KP455654) and Yolo Wonder (88.3%, JN019858) from the United States, Healey from Canada (99.1%, KT149366), IS from Israel (99.2%, JQ951943) and Lj from China (98.1%, KF709944). Isolate Kyosuzu (BPEV_KS) was shown to have 100% incidence in tested bell pepper cultivars in Japan and some of its variants were found to infect other C. annuum genotypes as well as related Capsicum species such as C. baccatum, C. chínense and C. frutescens (Okada et al., 2011). BPEV_KS can be transmitted through seed but not by graft inoculations and hybridization studies after denaturing agarose gel electro-phoresis detected a nick in the plus strand at position 880 (Okada et al., 2011). BPEV Yolo wonder (BPEV-YV) was the first BPEV to be sequenced (Okada et al., 2011). The other complete BPEV sequences have been only characterized in silico: isolate Maor was identified from transcript shotgun assembly as a contig with strong sequence identity to Bell pepper endornavirus (Jo et al., 2016); isolate Healey was identified in a NGS study of small RNAs extracted from the leaves of a pepper plant (cultivar Healey) with mild crinkling and chlorosis symptoms (Chen et al., 2015) and BPEV_IS was identified in asymptomatic C. annuum L. cv. Yatir leaves in Israel by NGS sequencing of viral small RNA (Sela et al., 2012). There are not published reports on isolate Lj from China. Outside the BPEV group, the closest endornavirus species is Hot pepper endornavirus (HPEV, KR080326) a proposed species (Lim et al., 2015) also infecting C. annuum that shares 78.8% nucleotide sequence identity with BPEV_Santa_Fe and BPEV_Peñol. Due to the high similarity between the Colombian BPEV isolates and BPEV_KS, it is likely for them to share the same molecular and biological properties.
A phylogenetic analysis using complete endornaviral proteins confirmed the previous analysis (Fig. 3A). As expected, BPEV_Santa_Fe and BPEV_Peñol clustered within the group of Bell pepper endornaviruses (bootstrap of 100%) with HPEV as a sister species. The Capsicum-infecting endornaviruses are part of a larger clade of plant endornaviruses that includes PvEV-2, infecting common bean and, more distantly, HvEV infecting barley. A comparison of functional domains reveals that BPEV, HPEV, PvEV2 and HvEV share methyltransferase, helicase and RNA-dependent RNA polymerase domains; however, HvEV lacks a UDP-glycosyltransferase present in the others (Fig. 3B). It has been suggested that the UDP-glycosyltransferase domain was transferred to endornaviruses from marine and freshwater bacteria (Song et al., 2013) and it is likely that this event probably occurred after the split between HvEV and the PvEV2/BPEV/HPEV group.
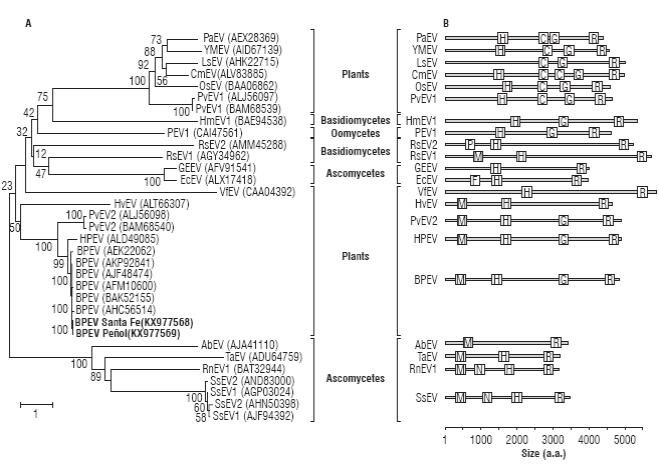
FIGURE 3 Evolutionary relationship of BPEV_Santa_Fe and BPEV Peñol to members of the genus Endornavirus. A) Neighbor-joining analysis of complete endornaviral polyproteins infecting plants, fungi (Basidiomycetes and Ascomycetes) and Oomycetes (Straminipila). The tree is drawn to scale with branch lengths in units of number of amino acids substitutions per site as indicated in the bar at the bottom. B) Relative location of functional domains of the polyproteins depicted in the tree on the left. Species abbreviations: Persea americana endornavirus, PaEV; Yerba mate endornavirus, YMEV; Lagenaria siceraria endornavirus, LsEV; Cucumis melo endornavirus, CmEV; Oryza sativa endornavirus, OsEV; Phaseolus vulgaris endornavirus 1, PvEV1; Helicobasidium mompa endornavirus, HmEV1; Phytophthora endornavirus 1, PEV1; Rhizoctonia solani endornavirus 2, RsEV2; Rhizoctonia cerealis endornavirus 1, RcEV1; Grapevine endophyte endornavirus 1, GEEV; Erysiphe cichoraceum endornavirus, EcEV; Vicia faba endornavirus, VfEV; Hordeum vulgare endornavirus, HvEV; Phaseolus vulgaris endornavirus 2, PvEV2; Hotpepper endornavirus, HPEV; Bell pepper endornavirus, BPEV; Alternaria brassicicola endornavirus, AbEV; Tuber aestivum endornavirus, TaEV; Rosellinia necatrix endornavirus 1, RnEV1; Sclerotinia sclerotiorum endornavirus 1; SsEV1; Sclerotinia sclerotiorum endornavirus 2; SsEV2. Domain abbreviations: putative methyltransferase, M; viral helicase, H; glycosyltransferase, G; RNA-dependent RNA-polymerase, R; Glycosyltransferase sugar-binding, C; Phospholipase A2, P; Furin-Like, F; Neuromodulin, N and DEAD-like helicase, D. Bootstrap values are shown above branches.
A global analysis of the phylogenetic relationships among endornaviruses, reveal two additional clades infecting plants. One of these clades has Vicia faba endornavirus (VfEV) as a sole member and, in spite of having the largest polyprotein, has only two identifiable domains with Helicase and RdRp function; VfEV is more closely related to fungal endornaviruses. The third group of plant endornaviruses comprises PvEV1, OsEV, CmEV, LsEV, YMEV and PaEV, characterized by the lack of a methyltransferase domain and the presence of an additional glycosyltransferase sugar binding domain containing a DXD motif (cl19952; Song et al., 2013). Interestingly, members of the OsEV group have been shown to localize in microsomal fractions while members of the BPEV group localize in the chloroplast, a fact that could be explained by their different domain structure (Moriyama et al., 1996; Okada et al., 2013). The polyprotein phylogenetic analysis confirms the complex evolutionary history of endornaviruses and suggests at least two independent origins of plant endornaviruses from fungi (Roossinck et al., 2011; Okada et al., 2013). It is likely that plant endornaviruses were first transferred to the plant kingdom from endornaviruses infecting ascomycetes fungi, all sharing Methyltransferase domains. Radiation of endornaviruses to basidiomycetes, oomycetes (Straminipila) and other ascomycetes seem to derive from a genetic transfer from plants, an evolutionary event characterized by the loss of the methyltransferase domain. A second transfer to plants from basidiomycetes fungi seem to have occurred which later resulted in the incorporation of a second Glycosyltransferase domain. It is interesting to note that all the hosts of non-plant endornaviruses have a close ecological relationship with plants as either pathogens (S. sclerotiorum, R. necatrix, A. brassicicola, E. cichoracearum, R. solani, R. cerealis, Phytophthora sp., H. longisporum) or soil-inhabitants (T. aestivum).
To confirm the widespread presence of endornaviruses infecting C. annumm cultivars in Antioquia primers BPEV_F, BPEV_R and qBPEV_R were designed using the sequenced genomes as reference (Fig. 4A). Primer BPEV_F binds to an internal segment of the RdRp domain which in combination with BPEV_R gives an amplification product of 909 bp useful in RT-PCR detection; in combination with primer qBPEV_R, BPEV_F amplifies a 100 bp segment suitable for RT-qPCR. Both primer sets were tested in C. annuum samples from different cultivation plots in Marinilla and El Peñol (Antioquia). In eight foliage samples, out of ten tested, RT-PCR gave amplification products with the expected size and sequence (Fig. 4B). Similar results were obtained by RT-qPCR using the BPEV_F/qBPEV_R primer set, in this case, sigmoidal amplification profiles were found in all the ten samples with Ct values in the 11.36-18.29 range (Fig. 4C). HRM analysis of the amplification reactions demonstrates that the amplification was specific, as evidenced by the presence of single denaturation peaks and their similar Tm values with respect to the NGS samples used as positive controls (Fig. 4D). Sanger sequencing confirmed the identity of the qRT-PCR amplicons. These results are in agreement with previous work suggesting a high incidence of BPEV in different Capsicum species such as C. annuum, C. frutescens and C. chinense (Okada et al., 2011; Sela et al., 2012; Chen et al., 2015; Jo et al., 2015; Lim et al., 2015; Jo et al., 2016). It has been shown the phylogenetic relationship between BPEV sequences mirror the evolutionary history of their hosts and it is likely that BPEV was present in the common ancestor giving rise to these three Capsicum species but not in primitive Capsicum species such as C. chacoense, C. annuum var. glabriusculum and C. pubescens (Okada et al., 2011). With Colombia being part of the center of origin of Capsicum species, it would be interesting that future investigations address the appearance of endonarviruses within this plant genus as well as their incidence in domesticated and wild species.
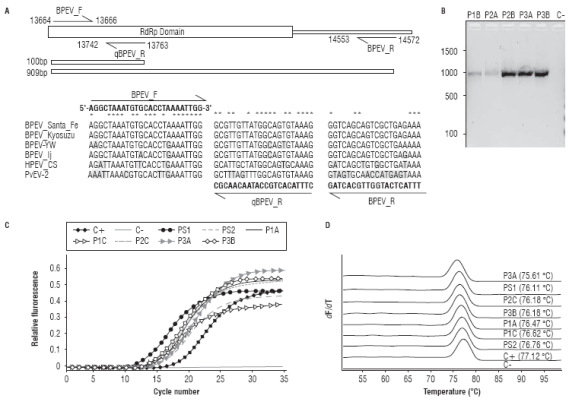
FIGURE 4 Molecular detection of BPEV in C. annuum samples from Marinilla and El Peñol. A. Location of the BPEV_F, BPEV_R and qBPEV R primers described in the text with respect to the BPEV_Santa_Fe consensus genome and the RdRp motif. The sequence alignment with selected endornaviral genomes highlights variable positions with respect to each primer. B. RT-PCR detection of BPEV using the BPEV_F/R primer set showing the expected amplification product of 909 bp in a subset of the field samples. C. Validation of the BPEV_F/ qBPEVR primer set in RT-qPCR. D. High Resolution Melting analysis of the amplification reactions shown in panel C. The positive control corresponds to the NGS sample sequenced from chili pepper.
Sequences derived from BPEV_F/BPEV_R amplicons (909 bp) were used to construct a phylogenetic tree with corresponding RdRp sequences of BPEV and another Capsicum-infecting endornaviruses like HPEV, with PvEV-2 as outgroup (Fig. 5). BPEV sequences form two clusters that correlated well with the infection host. BPEV group I comprises sequences infecting C. annuum cultivars and includes all the Colombian sequences reported in this work. Colombian sequences form a distinct group closely related to isolates Healey (Canada), Maor (USA), Kyosuzu (Japan) and Atir (Israel) and more distantly to isolate Lj from China. Two divergent BPEV sequences are observed within group I corresponding to isolates YW and LA-4 from the United States. BPEV group II comprises isolates LA-3 and LA-5 infecting yellow lantern chili (C. chinense) and isolates LA-1 and LA-2 infecting C. frutescens, a chili pepper species. Group II is more closely related to HPEV infecting C. annuum than to members of group I indicating that BPEV is a paraphyletic group as it does not include all sequences derived from the same common ancestor. A taxonomic revision by ICTV on the species comprising the BPEV clade is probably required.
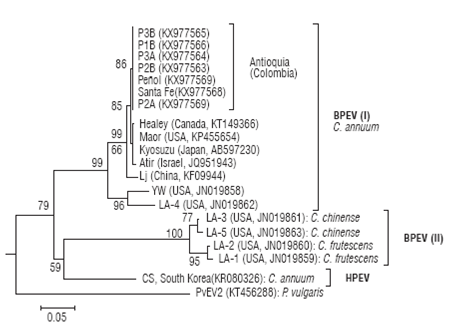
FIGURE 5 Evolutionary relationship between Bell pepper endornavirus isolates using partial nucleotide sequences of the RdRp región for a subset of the field samples. Sequences are labelled by the isolate name; location and GenBank accession codes are shown in parentheses. Phaseolus vulgaris endornavirus 2 was used as outgroup. The tree was constructed by the Neighbor-Joining method using distances calculated by the Tamura 3-parameter method. The tree is drawn to scale with branch lengths in units of number of base substitutions per site as indicated in the bar at the bottom. Bootstrap values are shown above the branches.
Conclusions
NGS sequencing of the C. annuum transcriptome revealed the presence of Bell pepper endornavirus (BPEV) in Antioquia (Colombia) closely related to isolates with worldwide distribution. These results were confirmed by sequencing of the RdRp region using primers BPEV_F/BPEV_R and by RT-qPCR using primers BPEV_F/qBPEV_R. The results presented here are in agreement with previous genomic studies on BPEV. Further work should aim at characterizing the molecular features of BPEV in different C. annuum cultivating regions in Colombia and confirm completion of the 5' and 3' ends.














