Introduction
Soybean (Glycine max) is a Fabaceae plant, whose seeds contain sugars (~30%), protein (~35%), edible oil (~20%), fiber, vitamins, and minerals. Soybean is a source of protein comparable to meat or eggs. Soybean cake is used as animal feed or industrial substrate (Widholm et al., 2010). The soybean production for 2016 was calculated at 320 million t. united States and Brazil are major soybean producer countries. United States has an estimated soybean production of 108 million t, and their harvested area is estimated at 33 million ha. Brazil has a harvested area of 33 million ha with a production of100 million t (USDA, 2017).
Colombia with an average production rate of 75,000 t in 2016 is considered a small soybean producer, occupying the 37th position worldwide, and the 6th in South America (Fenalce, 2017). Colombian soybean is developed mainly in the Eastern plains region, which is a key area for agricultural development due to its plain geography suitable for technification, vast land extensions, and development opportunities, especially in the post-conflict period. Considering that soybean is a plant originated and cultivated in temperate latitudes, and due that Colombia is a tropical country, several Colombian breeding programs have been developed since the 80s producing varieties adapted to local soil and weather conditions, including relevant differences in plant physiology parameters as the plant photoperiod and flowering processes that are correlated to the production rates (Valencia and Ligarreto, 2010). Currently, there are several soybean varieties with a great adaptation to local conditions, and high production (over 25,000 kg ha-1) which are used by local farmers.
The genetic modification by transgenesis (GM) in commercial plants has been a useful technology providing farmers with a tool to increase crop yield, reduce the quantity of pesticides, and increase the farmer profit (Klümper and Qaim, 2014). Soybean has been bred by transgenesis, including different traits as herbicide tolerance, insect pest resistance, and improvement of oil quality (ILSI research Foundation, 2017). Particularly, herbicide tolerance has been a very widespread and successful trait on soybean, having adoption rates of 94% in USA, 96.5% in Brazil, and 100% in Argentina in 2016 (ISAAA, 2016). GM soybean enhanced with herbicide tolerance has several advantages compared to the regular varieties. Among them, it allows the use of one herbicide per crop, a longer period of weed control, a lower Glyphosate concentration in soil, less herbicide application events, the use of low toxicity herbicides such as Glyphosate, and higher profits to the producer as an expression of all the above. Also, herbicide tolerant soybean is compositionally equivalent to the conventional genotypes and finally, the expiration of patents protecting herbicide tolerant soybeans can be a base for generic GM crops (Bonny, 2009). In Colombia, the government policies allow growers to produce transgenic plants and there are no restrictions to the consumers. Last reports indicate that in 2016 100,00o ha of GM maize were planted, of which 9,800 ha were GM cotton and 12 ha were GM flowers (ISAAA, 2016). Governments are committed to assess and manage the risks associated with the development and release of genetically modified crops. There is an established regulation of GM soybean specially attending the food safety affairs, including a maximum limit of herbicide, substantial equivalence, and varieties description. Countries have developed protocols by GM detection based on phenotypic or molecular assays (Tillmann et al., 2004).
Agrobacterium tumefaciens is a relevant microorganism used in transgenesis of crops. This process is based on the transfer of a DNA segment (T-DNA) from a tumor-inducing (Ti) plasmid, which is incorporated into the plant genome with its resultant expression. The T-DNA and Vir proteins form a molecular set that delivers a single strand of this T-DNA into the cell (Bourras et al., 2015). Soybean is a recalcitrant plant for Agrobacterium-mediated transformation producing low transformation efficiencies and requiring the use of hypervirulent bacterial strains and specific plant genotypes to allow its transformation (Atif et al., 2013). Some protocols have been applied to improve the transformation efficiency using A. tumefaciens; these methodologies include the modification of certain sanitation and infestation procedures toward observing possible differences among genotypes (Liu et al., 2013).
Glyphosate, whose chemical name is N-(phosphonomethyl) glycine (C3H8NO5p), is an odorless white strong acid. it is a crystalline powder with a fusion point of 184.5°C and molecular mass of 169.1 gmol-1. Glyphosate is a systemic herbicide and can be used with practically any type of crop to control weeds worldwide. Enzyme 5-enolpyruvyl-shikimate-3-phosphate synthase (EPSPS) is the target of Glyphosate, which is directly involved in the synthesis of the aromatic residues: phenylalanine, tyrosine, and tryptophan (Shikimate pathway). in 1996, the first GM soybean with Glyphosate tolerance was introduced in USA, expressing an EPSPS protein from a Cp4 strain of A. tumefaciens, which has no affinity for Glyphosate and allows the normal perform of the shikimate pathway (Duke and powles, 2008; Duke and Cerdeira, 2010). Glyphosate tolerance is a successful trait in soybean crop, and it currently offers the possibility to develop generic transgenic crops. Considering that patents that cover the development process of this particular trait recently expired (Jefferson et al., 2015), there is an increase in the freedom to operate (FTO) of the commercial plants related. Colombian soybean varieties have not been bred by transgenesis despite the fact that they are approved as commercial crops with Glyphosate tolerance (ICA, 2010) and that imported GM soybean seeds have low possibilities to grow well in tropical conditions. The main objective of this study was to evaluate the capacity of Colombian soybean varieties to be transformed by A. tumefaciens, using a Glyphosate tolerance expression cassette with FTO in Colombian territory, as a stage to produce and commercialize generic GM crops in Colombia.
Materials and methods
Plant genotypes
Three Colombian genotypes of soybean were used in transformation experiments: SK7 and p29 (bred by Kamerun and panorama companies respectively) and Soyica p-34 (bred by the Colombian Agricultural institute - ICA). These genotypes were chosen due to its high cultivation rate in the Eastern plains region. SK7 genotype is adapted to grow between 300 and 1,200 m a.s.l., a vegetative stage of 110112 d, height of 101 cm, white flowers, brown pubescence, oil content of 20.47%, and a yield rate of 2,600 kg ha-1. p29 genotype is adapted to grow between 300 and 1,200 m a.s.l. as well, has a vegetative stage of 105-115 d, height of 100 cm, purple flowers, brown pubescence, an oil content of 21.37%, and a yield rate of 2,600 kg ha-1. Soyica p-34 is equally adapted to the same altitude (300 to 1,200 m a.s.l.), a vegetative stage of 110 d, height of 69 cm, white flowers, brown pubescence, an oil content of 20%, and a yield rate of 2,700 kg ha-1. As start material field conditions seed were used, without any fungicidal or insecticidal treatment, and a moisture between 11 to 12%. These varieties were chosen because they were the most demanded by farmers at the time in which the experiment was performed.
Agrobacterium strains, expression cassette and vector
A previously reported cassette designated as E-IGP was used; this cassette contains a polyubiquitin promoter from soybean (GmUbi), followed by a transit peptide from Petunia hybrida, a codon optimized cp4 epsps gene for expression in soybean tissues, and a nopaline synthase (NOS) terminator codon (Jiménez, 2014). The E-IGP cassette was introduced into a pCAMBiA1301 vector on which the GUS reporter gene and the hygromycin selection gene were excised in such a way that only the E-IGP cassette could be transferred to the plant genome (Jiménez, 2014). The vector containing the E-IGP cassette was introduced into A. tumefaciens strains AGLO (bought from an American Type Culture Collection, under ATCC®BAA-1OOTM denomination) and EHA1O5 (acquired by a donation from the Cenicaña institution). Recombinant strains were maintained in Luria Bertani (LB) medium containing 5O mg L-1 kanamycin. Agrobacterium cultures used for infection of explants were grown in LB medium. The observed optical density at 65O nm (OD65O) ranged from O.8 to 1.O, at 28°C and 2OO rpm. A bacterial pellet was obtained by centrifugation of 3O ml of bacterial culture at 8OOO rpm for 4 min at 2O°C. The pellet was resuspended in 25 ml of co-cultivation liquid medium (CCLM) (1X Gamborg vitamins, O.1X B5 salts (Gamborg et al., 1968), 1.67 mg L-1 benzylaminopurine (BAP), O.25 mg L-1 gibberellic acid (GA3), 3% sucrose, 2O mM2-[N-Morpholino] ethanesulfonic acid (MES), 2OO μ macetosyringone, pH 5.7) and then was used as inoculum for the plant tissues.
Explant preparation and A. tumefaciens infection
Soybean seeds were selected considering their appearance, choosing those that did not have lacerations or spots, and had a homogeneous size. The seeds surface was sterilized following the chlorine gas technique (Paz et al., 2OO4; Paz et al., 2OO6; Song et al., 2O13) for 16 h, generating gas from a mix of 4.1 ml 1O N HCl with 1OO ml 5% NaClO. Sterilized seeds were germinated on 0.7% plant Tissue Culture (PTC) agar medium (water plus agar), pH 5.7, with the hilum proximal to the media for 5 d, and incubated under a 16/8 (light/dark) photoperiod at 26°C.
Once seeds were germinated, the seed coat was eliminated and a cutting was done 5 mm below the cotyledons junction to eliminate the hypocotyl. After that, cotyledons were separated by a longitudinal cut on the remaining piece of hypocotyl. Plumule was eliminated from both cotyledons, and some incisions (7-12) were made on the cotyledonary node. Each cotyledon with its own cotyledonary node was considered as an explant to transformation. Explants were infected with A. tumefaciens strains by submerging them in a CCLM solution containing bacterial biomass for 3O min, followed by cultivation on a co-cultivation medium (CCM) (CCLM added with 0.7% PTC agar), with the ad-axial side down, and incubated in the dark for 3 d at 28°C (Zhang et al., 1999).
Regeneration test of soybean varieties
A first trial was performed to observe the behavior of soybean varieties in an in vitro system intended for Agrobacterium-mediated transformation. For such task, a regeneration ability assay was carried out for every variety without Agrobacterium infection. In this essay, 2OO seeds of each variety were selected and germinated obtaining 24O explants that were prepared as described above. These explants were cultivated on CCM without Agrobacterium strain for 3 d, and later were cultivated on a shoot induction medium (SIM) (1X B5 salts, 1X Gamborg vitamins (Gamborg et al., 1968), 3% sucrose, 1,67 mg L-1 BAP, 3mM MES, O.7% PTC agar, pH 5.7) for 4 weeks, with a medium replacement at the end of the second week.
After four weeks on SIM, the number of explants producing at least one shoot (regenerating explants) was recorded as well as the number of shoots produced by each regenerating explant.
Transformation, regeneration and selection
Transformation assays were performed following the methodology described by Zhang et al. (1999), plus some modifications. To assess the ability of each vegetal variety to be transformed with E-IGP cassette, the assay was divided in two treatments and two controls as follows: Treatment 1: three varieties transformed with an EHA1O5 strain containing a E-IGP cassette, and selected in vitro using the herbicide Glyphosate; Treatment 2: three varieties transformed with an AGLO strain containing a E-IGP cassette, and selected in vitro using the herbicide Glyphosate; Control 1 (relative control): three varieties without transformation, and selected in vitro using Glyphosate; and Control 2 (absolute control): three varieties without transformation, and without in vitro selection.
For each treatment and control, 13O explants of each variety as described above were prepared. Treatments were inoculated with Agrobacterium strains and co-cultivated as described above. Controls were cultivated on CCM in the same way as treatments, but without bacteria. After co-cultivation, all explants were rinsed in sterile water added with 5O mg L-1 cabenicillin, on a rotary shaker at 41O rpm for 4O min, three times. After this rinse, the explants were transferred to SIM mixed with antibiotics (25O mg L-1 cefotaxime, 1OO mg L-1 timentin), and they were incubated for two weeks, under a 16/8 (light/dark) photoperiod at 26°C. After this procedure, SIM was renewed with a fresh SIM plus antibiotics solution and mixed with a 148 μM Glyphosate reagent grade (phytotechnology Laboratories®, Lenexa, KS, USA). This methodology was followed in Treatments 1, 2 and in Control 1; Glyphosate was not added in Control 2. During the process to transfer explants to fresh SIM, the remaining hypocotyl of each explant was cut to allow the fresh tissue to directly contact the growth medium. Growing process was performed for two additional weeks under the same conditions.
Once the shoot induction period was finished, it was followed by a shoot elongation period (SEP). All explants were cut to eliminate remaining cotyledon and thus allowing fresh tissue to be in contact with the medium. Explants developing at least one shoot were transferred to Shoot Elongation Medium (SEM) (1X B5 salts, 1X Gamborg vitamins (Gamborg et al., 1968), 3% sucrose, O.5 mg L4 GA3, O.1 mg L-1 indole acetic acid (IAA), O.7 mg L-1 BAP, 5O mg L-1 glutamine, 5O mg L-1 asparagine, 3 mM MES, 25O mg L-1 cefotaxime, 1OO mg L-1 timentin, 0.7% PTC agar, pH 5.7) added with a 35 μM Glyphosate reagent grade (phytotech-nology Laboratories®, Lenexa, KS, USA) in Treatments 1, 2 and in Control 1, and without Glyphosate in Control 2. The SEM solution was replaced in the explants every two weeks for fresh SEM. In treatments and control containing Glyphosate, the herbicide was added during four weeks, removing it between the fifth week and until the end of SER The explants were allowed to grow until shoots reached a height of 3 cm, for a maximum SEP of 1O weeks, under 16/8 (light/dark) photoperiod to 26°C.
Shoots that reached the required height (3 cm) were individualized, labeled as "KJ" for SK7 variety, ״PJ" for p29 variety and "SJ" for Soyica p34 variety, and marked by a number to indicate a consecutive individualization, thus discriminating the lines obtained from different treatments. Each individual line was transferred to a propagation Medium (PM) (MS salts O.66X (Murashige and Skoog, 1962), (1X Gamborg vitamins (Gamborg et al., 1968), 3% sucrose, 0.7% PTC agar, pH 5.7), and it was propagated to obtain biomass to consume in molecular and phenotypical analysis.
Molecular and phenotypical analysis
Individualized lines that were successfully propagated were subjected to the polymerase chain reaction (PCR) analysis to detect transgene insertion. Genomic DNA was extracted from each line following CTAB buffer methodology (Doyle, 1991), and it was quantified spectrophotometrically using NanoDrop equipment (Thermo Fisher Scientific, Waltham, MA USA). A 2O5 bp region of transgene was amplified using the pair of primers 5'-CTTTGCTGAA-GGAGCTACCG-3' and 5'-GTGATCGAGATGCGTAG-CAA-3' along with reagents included in Kapa3G plant PCR kit (Sigma-Aldrich Corp. St. Louis, MO, USA) following manufacturer's instructions. PCR products were separated by electrophoresis on a 1% agarose gel. Transformation frequency was calculated as number of positive PCR lines / total number of transformed explants χ 100.
To detect EPSPS expression, the vegetal biomass of propagated lines was subjected to the commercial Enzyme Linked Immunosorbent Assay (ELISA) Roundup Ready Cp4 EPSPS (Agdia Inc. Elkhart, IN, USA), following manufacturer's instructions. Absorbance of each sample was measured using iMarkTM Microplate Reader (Biorad, Hercules, CA, USA) at 655 nm wavelength. percentage of functional transformants was calculated as number of positive ELISA lines / total number of transformed explants χ 100.
On each positive PCR sample, a Southern Blotting assay was performed following recommendations of digoxigenin (DIG) applications platform for filter hybridization (Sigma-Aldrich Corp. St. Louis, MO, USA) (Eisel et al, 2008). A DIG-labeled probe was synthesized using the PCR DIG probe Synthesis kit (Sigma-Aldrich Corp., St. Louis, MO, USA) with the same primers described above for PCR. Twenty μg of genomic DNA were digested with PvuII and NdeI enzymes (New England Biolabs® Inc., Ipswich, MS, USA) in a double digest reaction. Restriction fragments were separated on 0.7% agarose gel by electrophoresis and transferred onto a positively charged nylon membrane (Sigma-Aldrich Corp., St. Louis, MO, USA). prehybridization, hybridization, membrane washing, and detection were conducted following the platform instruction manual (Eisel et al., 2008). The detection of probe-target hybrids was done by chemiluminescence using CDP-Star substrate (Sigma-Aldrich Corp., St. Louis, MO, USA).
Only a small amount of possible transgenic lines were allowed to grow on soil-type substrate. On hardened lines, a "plant painting" was performed using a 0.5% dilution of commercial Glyphosate (Victorius® 48 SL, Sodiak SA, Bogota, Colombia), over the shoot of the plant. Herbicide affectation was recorded after 10 d based on plant survival. parallel to application on transformed lines, the same Glyphosate solution (0.5%) was applied on a non-transformed plant as positive control for test herbicide activity. In contrast, solvent (tap water) was applied on another non-transformed plant as a negative control.
Results
Regeneration capacity of soybean varieties
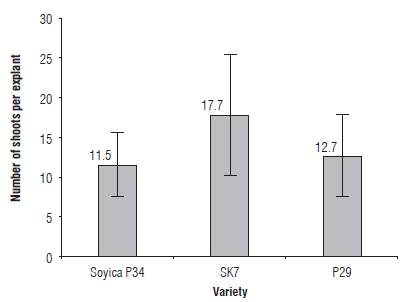
Three Colombian soybean varieties were evaluated regarding their regeneration capacity as a preliminary clue of how their behavior on an Agrobacterium-mediated transformation system will be. Starting from 240 explants of each variety, 212 explants of Soyica p34 (88.3%), 181 explants of SK7 (75.4%) and 231 explants of p29 (96.2%) developed regenerated shoots. After counting the number of shoots per explant (on regenerative explants) in each variety, it was found that, on average, Soyica p34 regenerates 11.5 shoots per explant (± 3.9 shoots), SK7 regenerates 17.7 shoots per explant (± 7.73 shoots) and p29 regenerates 12.7 shoots per explant (± 5.2 shoots) (Fig.1).
At first the non-parametric Kruskal-Wallis test was applied using R software, to compare if varieties outputs have equality in the middle range or if at least one of them is different (Hollander et al., 2014). In statistical terms, some of the following hypothesis systems were assessed:
When the test was carried out, a P-value of O was obtained, and the null hypothesis was rejected, suggesting that at least one of these average ranges is different from the others. Therefore, the non-parametric Wilcoxon test was used, in order to compare the differences on the average ranges of two samples, and to identify which is higher (Hollander et al., 2014). The test was carried out two by two, with α = 0.1, as follows:
The p-values associated with the tests performed were 0.00 for H1, 0.99 for H2 and 0.00 for H3. For any p-value smaller than α, the null hypothesis is rejected, indicating that the mean range of shoots of the SK7 variety is greater than the mean range of the Soyica P34 and p29 varieties.
Agrobacterium-mediated transformation
The three Colombian soybean varieties described above were subjected to a genetic modification. Such methodology consisted in an insertion of a transgene designed to express a Cp4 EPSPS protein to confer tolerance to the herbicide Glyphosate. This process was performed using the A. tumefaciens bacteria as a transgene's vehicle, assessing two bacterial strains, AGLO and EHA105.
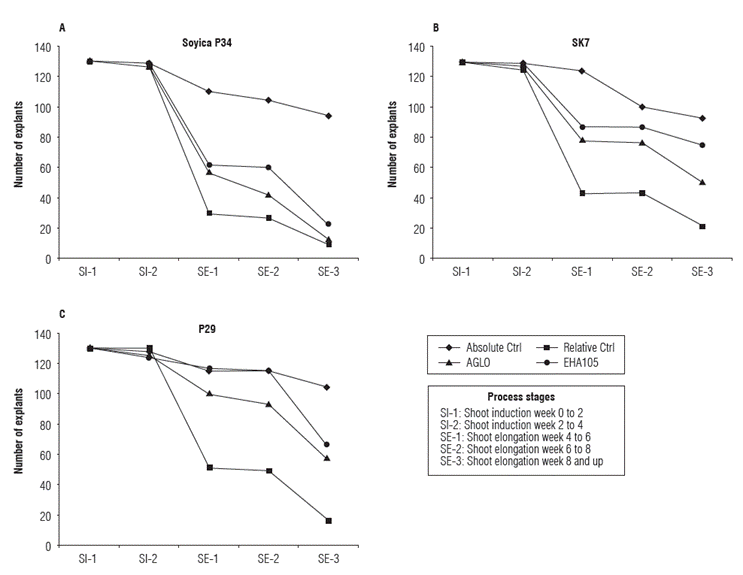
To assess the behavior of treatments and controls in each variety, the explant number in each process stage prior line individualization was recorded. Each treatment and control had initially 130 explants, and this number was decreasing in successive stages (Fig. 2). It was observed that in all three plant varieties, addition of Glyphosate to the culture medium reduced the explants regeneration compared to the control without herbicide addition. Taking as a reference the explants subjected to absolute control that reached the shoot elongation 3 stage (SE-3) in each variety (100%), it was observed that the highest damage by herbicide was presented in Soyica p34. For this variety, 8.5% of explants in relative control, 12.8% in transformation with strain AGLO and 22.3% in transformation with strain EHA1O5 reached the SE-3 stage. Furthermore, for p29 16% of explants in relative control, 55.8% in transformation with strain AGLO and 64.4% in transformation with strain EHA105 reached the SE-3 stage. The most tolerant variety to herbicide addition was SK7, in which 23.7% of explants in relative control, 54.8% in transformation with AGLO and 80.6% in transformation with strain EHA105 reached SE-3 stage (Fig. 2).
After reaching the SE-3 stage, the shoots that grew above 3 cm were labeled and marked. These Lines were propagated, and only those that were successful were subjected to molecular and phenotypical analysis. A total of 91 possible primary transformants of all varieties and treatments were labeled (Tab. 1), but only 53 lines were effectively propagated, so DNA extraction was performed exclusively on those 53 lines. PCR was done on extracted DNA and separated by electrophoresis (Fig. 3).
TABLE 1 Results of transgene presence / absence test and CP4 EPSPS protein detection on generated lines.
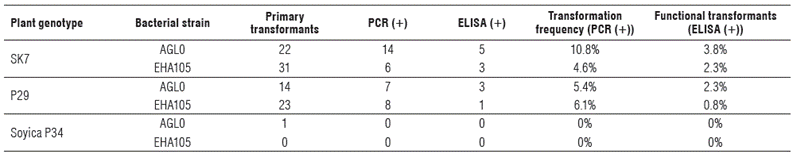
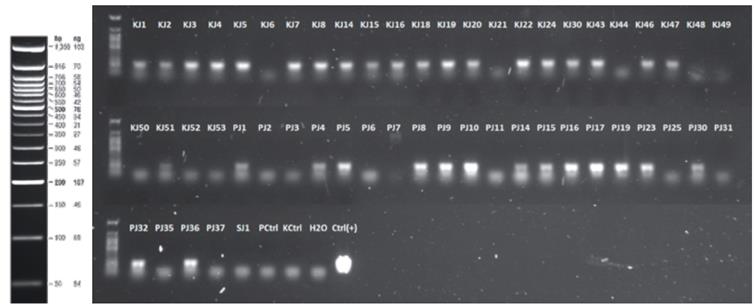
FIGURE 3 Detection of the E-IGP cassette on possible primary transformants. The name of each line is indicated at the top of each lane. PCtrl: Non transformed P29 variety, KCtrl: Non transformed SK7 variety, H2O: absolute control with primers, Ctrl (+): positive control, corresponding to plas-mid vector E-IGP extracted by miniprep. The molecular weight marker corresponds to 50 bp DNA Ladder (New England Biolabs).
SK7 showed the highest transformation frequency (10.8%) when it was combined with AGLO strain (Tab. 1). p29 was also efficiently transformed in a lower proportion, (5.4%) when it was combined with AGLO strain, and 6.1% when it was combined with EHA1O5 strain. It was not possible to transform Soyica P34 variety under the procedures followed in this research (Tab. 1).
To assess the protein expression, an ELISA test on biomass of the propagated plants from possible primary transformants was performed. In this test positive results by 12 lines of SK7 and P29 varieties were obtained (Tab. 1). Thus, the highest percentage of functional transformants was obtained for SK7 when it was combined with AGLO (3.8%). For p29 variety, the highest percentage of functional transformants was obtained also by using AGLO strain (2.3%) (Tab. 1).
A Southern blot was performed on positive PCR samples. All of them showed a single insertion band, ranging between 6.5 to 24 kbp at a different size in every DNA sample (Fig. 4).
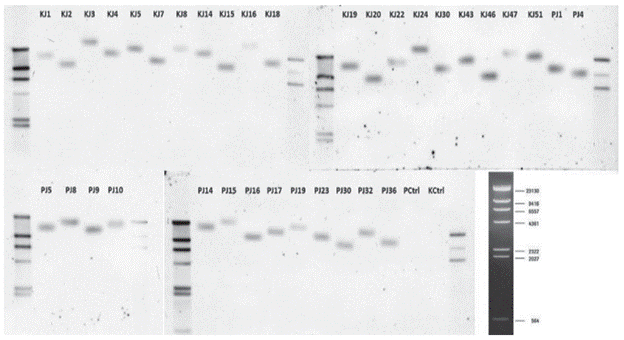
FIGURE 4 Southern blotting on positive PCR samples. The name of each transformant is indicated on the top of each lane. PCtrl: Non transformed P29 variety, KCtrl: Non transformed SK7 variety. Each membrane has, on its left side, a molecular marker, corresponding to DNA Molecular Weight Marker II, DIG-labeled (Sigma-Aldrich Corp. St. Louis, MO, USA), and, on its right side, a positive control, corresponding to E-IGP cassette integrated into pCAMBIA1301 non-linearized vector.
Some lines allow their hardening on soil-type substrate, supplying material to perform some greenhouse phenotypical tests. After applying a 0.5% solution of the commercial Glyphosate dose in a time period of 10 d, tolerance was assumed as the survival of treated plants. In contrast, susceptibility was considered as death of treated plants. The KJ7, KJ8, KJ15, KJ18 and PJ32 lines were found to effectively tolerate the herbicide, as there was no plant death after 10 d (Fig. 5C), whereas lines KJ24, KJ47 and SJ1 did not tolerate the herbicide application and died (Fig. 5D).

FIGURE 5 Greenhouse test for Glyphosate tolerance. A. Negative control, corresponding to a non-transformed plant painted with the herbicide solvent (tap water). B. Positive control, corresponding to a non-transformed plant painted with 0.5% commercial Glyphosate. C. Tolerant line, corresponding to KJ7 line. D. Susceptible line, corresponding to SJ1 line. C and D were painted with 0.5% commercial Glyphosate. Labels regarding the name of the lines are located at the top of each picture. The next day after the application event is located at the bottom of each picture.
Discussion
Transformation efficiency in soybean using A. tumefaciens is, in general terms low, mainly due to difficulties in the T-DNA transfer efficiency. To obtain transformed plants in regeneration systems, it is necessary to use hypervirulent strains of Agrobacterium (Atif et al., 2013). In the present study, three soybean varieties adapted to cropping conditions in Colombian territory were used, and considering that it is the first research carried out in the country on soybean genetic transformation, it was absolutely necessary to establish a transformation platform for this species, evaluating diverse factors that could affect the process, as the predisposition of local varieties to be genetically transformed.
The transformation efficiency in soybean is highly genotype-dependent (Atif et al., 2013), which may be linked to its regeneration capacity, among other factors. Paz et al. (2004) reported that Williams variety had a higher rate of regeneration within a set of 10 different soybean varieties with 100% regeneration. Within this set, the lowest rate of regeneration was obtained by the Harosoy variety, with 68%. In this study, the selected plant genotypes had different performances in each of the developed tests. In evaluation of regeneration capacity, it was observed that the p29 variety had the highest percentage of regeneration (96.2%), while SK7 had the lowest percentage (75.4%).
However, SK7 compensates its low regeneration percentage with a high rate of shoot production per explant, with 17.7 shoots on average, being the highest value among the evaluated genotypes. In general, regeneration of the three plant varieties was good with the tested system, considering that percentage values of explants with shoots and number of shoots per explant are adequate to take as baseline on a process of genetic transformation.
Regeneration differences between genotypes could be related to the balance of growth regulators present in Sim and its interaction with tissues and endogenous factors of each variety. Concentration of growth regulators must be optimized for each variety separately (Paz et al., 2006; Soto et al., 2013; Li et al., 2017).
It is important to include a selector agent in SIM to avoid the generation of chimerisms, or escapes from the transformation system. Most soybean transformation trials follow an in-vitro and greenhouse selection using glufosinate ammonium (Zhang et al., 1999; Paz et al., 2004; Zeng et al., 2004; Paz et al., 2006; Xue et al., 2006; Song et al., 2013; Jia et al., 2015; Yang et al., 2016; Li et al., 2017), Kanamycin (Liu et al., 2004) or Hygromycin (Olhoft et al., 2003; Arun et al., 2015; Kuma et al., 2015). There are fewer reports on Glyphosate selection (Clemente et al., 2000; Soto et al., 2016). In this report, a selection scheme was used to produce adequate plants further individualized and propagated as primary transformants. It is important to emphasize that Glyphosate selection is based on phenotypical criteria (shoot height and appearance) instead of the categorical criteria used for other selector agents as live/dead shoots.
The maximum transformation frequencies obtained in other reports, with different soybean varieties, are generally ranging between 4 and 15.8%, with an average of 8.5% (Olhoft et al., 2003; Liu et al., 2004; Paz et al., 2004; Zeng et al., 2004; paz et al., 2006; Yukawa et al., 2008; Jia et al., 2015; Yang et al., 2016; Li et al., 2017). In this report, a maximum transformation frequency on SK7 variety of 10.8% was obtained, which is above the average, reported for other varieties. P29 variety was also transformed with a frequency of 6.1%. This indicates that there are Colombian soybean varieties with potential to be genetically transformed, in this case, with special success on SK7 variety which had the highest regeneration capacity and also the highest transformation frequency.
Differences between varieties regarding their capacity to be transformed can be associated with defensive responses of plant tissues against bacterial infection and the capacity of cellular division, which varies between plant genotypes (Jia et al., 2015). There was a slight difference between both bacterial strains in relation to their infective capacity, showing AGL0 a higher infective ability than EHA105 on SK7. Both strains are derivatives from EHA101 strain, which in turn is derived from hypervirulent A281 strain, with a C58 chromosomic background (a nopaline type strain) (Lazo et al., 1991; Hood et al., 1993). So, considering that both strains have the same chromosomic background and vir helper vector, similar results are expected.
Recently, genome editing has been developed by addition, removal, or alteration on specific bases in the plant genome. a relevant technology of genome edition is CNSPR-Cas9 (clustered regularly interspaced short palindromic repeats associated to protein Cas9). The CNSPR-Cas9 technology has their origin in a bacterial defense system from viruses using RNA segments to target viral DNA, so as a result, Cas9 protein cut the parasite DNA. Basically, CNSPR-Cas9 is based on a guide RNA segment that binds to a specific sequence of DNA, using a Cas9 enzyme. This enzyme cuts the target DNA and repairs the cell machinery to add or remove the desired sequences. CNSPR-Cas9 system leaves no traces on the genome, which has been postulated as a form of genome modification different to transgenesis (Hussain et al., 2018). This affair was assessed by the European union on july 25, 2018 claiming that genome edition should be subject to the same regulations as the conventional GM crops. CNSPR-Cas9 system has been applied in soybean by stress tolerance (edition of Sucrose non-fermenting related protein kinases), multiple loci edition, flowering delay (by edition of GmFT2a gene in a specific base or short deletion), and promoter edition with relevance in expression, among others (Du et al., 2016; Cai et al., 2018; Li et al., 2018; Kanazashi et al., 2018).
This work is a preliminary approach to establish a stable transformation platform for crops of economic relevance such as soybean in Colombia. It was possible to produce entire plants with evident Glyphosate tolerance in greenhouse conditions, but the process should be subjected to further optimization to get a totally refined platform. This is an important step to initiate the production of generic transgenics in this country, specifically designed to develop Glyphosate tolerance as a transgenic trait in soybean varieties, while the patents protecting this technology expired in recent years (Jefferson et al, 2015).