Introduction
The chili pepper (Capsicum spp. L.) is one of the most important vegetables worldwide because of its culinary and medicinal uses (Green and Kim, 1991). Currently, the region of Valle del Cauca is one of the largest producers of chili pepper in Colombia, due to the climatic conditions and nutritional characteristics of the soil. However, the yield has decreased in the past ten years (Ministerio de Agricultura y Desarrollo Rural, 2018) as a result of factors such as temperature and humidity that are constantly changing due to climate change or diseases related to fungal, bacterial and viral pathogens; this last factor has become a major limitation for chili pepper production worldwide (Moury and Verdin, 2012).
In the tropics, viral diseases affect crop production and are common in plants of the Solanaceae family like chili pepper. These viruses belonging mainly to genera Begomovirus, Potyvirus, Tospovirus and Cucumovirus and affect chili pepper crops, causing more than 90% losses (Subramanya-Sastry, 2013). Although a great variety of viral genera have been reported as infectious agents, few studies have been carried out in the Valle del Cauca region or in Colombia.
Only the presence of a potyvirus, the pepper deforming mosaic virus (PepDMV) (Morales et al., 2005) and recently, a begomovirus (Vaca-Vaca et al., 2019) have been reported as presently affecting these crops. The cucumber mosaic virus (CMV) has only been detected using serological techniques like ELISA. However, these techniques have been displaced by molecular strategies that are more sensitive (González-Garza, 2017).
In Colombia, since 2005 no studies of RNA viruses in chili pepper have been published, so there is no accurate information on the current viral diseases that affect chili pepper crops in this biogeographic area. Therefore, it is likely that due to current global climate changes, a variation in the dynamics of populations of the viral vectors (insects) has occurred, causing changes in virus populations. So, the scenario of those last studies does not match the current situation of the dynamics of RNA viruses. This is more evident if we consider that these viruses evolve following the "Quasispecies" model (Shukla et al., 1994) which, through high mutation rates can generate new variants and in most cases new viral species that could emerge as new and more aggressive viral pathotypes.
CMV has a genome composed of three segments of positive single-stranded RNA (RNA1, RNA2 and RNA3). Each viral RNA fragment is encapsidated in an isometric particle and translated into important proteins responsible for the encapsidation, replication, and viral movement through the plant (International Committee on Taxonomy of Viruses, 2018). This virus is transmitted by aphids in a non-persistent manner (Scott, 2006). CMV isolates have been clustered into two groups, I and II, according to serology, nucleic acid hybridization, and gene sequences (Owen and Palukaitis, 1988; Wahyuni et al, 1992; Dubey et al., 2008). Then, Roossinck et al. (1999) divided subgroup I into IA and IB based on gene sequences and phylogenetic analysis. CMV is one of the major viral diseases worldwide due to its global distribution as well as its host range covering more than 1000 plant species (International Committee on Taxonomy of Viruses, 2018). In chili pepper crops, CMV produces some symptoms like mosaics, leaf distortion, stunted growth and fruit lesions, which result in a reduction in yield (Rahman et al., 2016; Azizan et al, 2017).
The aim of this research was the detection and molecular characterization of CMV in three different chili pepper varieties in the Valle del Cauca region of Colombia. Based on this information, the producers may be able to develop strategies to control this virus as well as having the necessary information to implement improvement programs for controlling viral diseases.
Materials and methods
Plant material
Seventy-one samples of three chili pepper varieties, habanero (Capsicum chinense), tabasco (Capsicum frutescens) and cayenne (Capsicum annuum var. acuminatum), at different phenological stages and with viral symptomatology were randomly collected in four municipalities (Palmira, Yumbo, Vijes and Yotoco) of Valle del Cauca in Colombia. Tissues collected from each plant were ground using liquid nitrogen and stored in a refrigerator at -20°C for further analysis.
RNA extraction
Total RNA extraction was performed from 100 mg of frozen or dehydrated plant tissue (with silica gel), using the TRIZOL® reagent (Invitrogen™). The quality and quantity of total RNA was verified in 0.7% agarose gels stained with ethidium bromide. These gels were visualized in a Chemi-Doc™ XRS photodocument, using the Quantity One 4.6 software (BioRad™, California, USA).
Virus detection by RT-PCR
For cDNA synthesis, 5 of total RNA was used as a template using the RevertAid First Strand cDNA Synthesis kit (ThermoFisher Scientific™, Massachusetts, USA). The cDNA was synthesized using both primers included in the kit: random hexamer and oligo dT. In order to detect CMV, a 586 bp fragment corresponding to the gene coding for the capsid protein (CP) was amplified by RT-PCR (Herrera-Vasquez et al, 2009) using the BIOLASE™ PCR kit DNA Polymerase (Bioline™, London, UK). For this reaction, a primer melting temperature (Tm) of 50°C was used. The amplification of the fragment was carried out in a C100 thermal cycler (BioRad™, California, USA). In order to characterize the CMV detected in chili pepper, the 586 bp fragment was purified using the Wizard® DNA Clean up system (Promega®, Wisconsin, USA) purification kit and was sent to Macrogen® (Seoul, South Korea) for Sanger sequencing.
Bioinformatics analysis
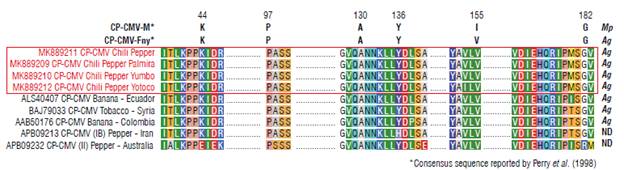
In order to know the identity of the 586 bp fragment, a sequence nucleotide analysis was performed by BlastN algorithm using the default search algorithm parameters: database: refseq genomes, organism: virus, program selection: somewhat similar sequences (blastn). The sequence was edited using the BioEdit program (North Carolina, USA); a multiple alignment was performed in the Clustal W program (Larkin et al., 2007). The identity values between the sequences, at nucleotide level, were calculated using the Sequence Demarcation Tool (SDT) 1.2 program (Muhire et al., 2014). Phylogenetic analysis was performed using the Mega 7 program (Kumar et al., 2016). The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987) and evolutionary distances were computed using the maximum composite likelihood method (Tamura et al., 2004). Also, a protein alignment was carried out to manually identify the key amino acids for vector transmission (Perry et al., 1998).
Design of specific primers for CMV chili pepper detection
Based on the CMV sequence obtained, a new set of primers was designed using the CLC main workbench v7.0® program (Qiagen®, Hilden, Germany). These primers amplified a 229 bp fragment of the CP gene. Then, the amplification conditions were standardized by gradient PCR. To evaluate the specificity of the primers, cDNA from different plants, including the three chili pepper varieties infected with this virus, were used as a template.
Mechanical inoculation of CMV chili pepper
Using inoculum from the field (tabasco chili pepper plants infected only with CMV), mechanical inoculation of CMV into 15 healthy and young (one month) plants of tabasco chili pepper (Capsicum frutescens) was performed under greenhouse conditions. Carburundum powder (600 mesh) and phosphate buffer pH: 7.4 (Na2HPO4 + NaH2PO4) were used for this purpose. One month after inoculation, CMV was detected by RT-PCR using the specific primers that we previously designed (CMV chili pepper-F / CMV chili pepper-R).
Results and discussion
Detection by RT-PCR of cucumber mosaic virus (CMV) in chili pepper
Ten samples of habanero pepper (Capsicum chinense), 59 samples of tabasco pepper (Capsicum frutescens), and two samples of cayenne pepper (Capsicum annuum var. acuminatum) were collected in 2017. The collected samples showed symptoms related to the viral disease such as leaf deformation and mosaics (Fig. 1).

FIGURE 1 Tabasco pepper with viral symptomatology: mosaics and severe leaf deformation, A) Palmira; B) Vijes: mosaic and mild leaf deformation; C) Yumbo; D) Yotoco.
Detection of CMV using primers described by Herrera-Vásquez et al. (2009) was carried out and fragments of the expected size (~586 bp) in 52.1% of analyzed samples (37/71) were obtained: 100% (6/6) of the samples from Palmira, 50% (12/24) of the samples from Yumbo, 56.5% (17/30) of the samples from Vijes and 27.2% (3/11) of the samples from Yotoco (Fig. 2). The highest number of positive samples for CMV was found in Palmira and Vijes, where severe viral symptoms were observed, such as the presence of mosaics and leaf deformation (Fig. 1A-B). The samples collected in Yotoco had a lower viral presence, reflected in the presence of mild symptoms (Fig. 1D).
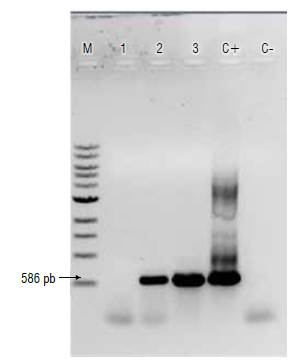
FIGURE 2 Detection of cucumber mosaic virus by RT-PCR in chili pepper. Products were analyzed through electrophoresis on 1% agarose gel. M: 1 kb DNA ladder; lane 1: A209 (Vijes), lane 2: A150 (Palmira), lane 3: A239 (Yotoco), C+: positive control (Plasmid DNA containing a CMV fragment), C-: negative control (water). The arrow indicates the expected fragment size.
Similar results have been found in other chili pepper varieties. Valadez et al. (2019) detected CMV by means of RT-PCR in jalapeño pepper in the Colima region of Mexico. Robles-Hernández et al. (2010) describe that symptoms, such as mosaics and leaf distortion, necrosis, defoliation, and flower fall, are known in jalapeño pepper crops in Mexico. These symptoms result in the decrease of fruits and can generate almost 90% of pepper crop losses.
Molecular characterization of CMV chili pepper
In order to confirm the identity of the detected virus in chili pepper crops, two CMV fragments from samples collected in each municipality were sequenced for a total of eight CMV fragments. A sequence of 549 bp of the CP gene was obtained for each fragment and an identity analysis was carried out among all the sequences of this study. First of all, both sequences from each municipality were the same.
Then, a 98.5% to 100% identity was found among all the sequences: the quasispecies from Palmira and Yumbo are 100% identical, and at the same time, they are 99.45% identical to the quasispecies from Vijes and 98.54% identical to the quasispecies from Yotoco. Finally, the quasispecies of Vijes and Yotoco share 99% identity. This might indicate that the CMV quasispecies found in Palmira, Yumbo, Vijes and Yotoco are the same.
Therefore, a sequence from each municipality was chosen for subsequent analysis. The sequence of these fragments was uploaded to the GenBank database with the following accession numbers: CMV-chili pepper isolate Palmira: MK889209, CMV- chili pepper isolate Yumbo: MK889210, CMV-chili pepper isolate Vijes: MK889211, and CMV-chili pepper isolate Yotoco: MK889212.
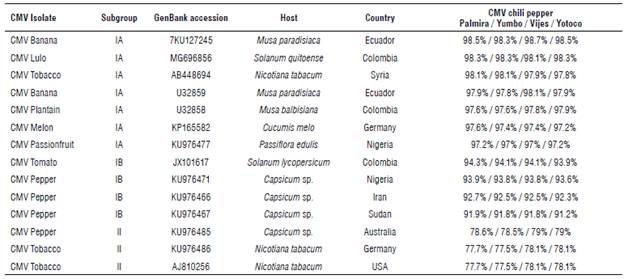
In order to know the percentage of identity of our quasispecies with other isolates from the GenBank database, an identity analysis was carried out in SDT (Tab. 1). Our CMV quasispecies isolated from chili pepper are highly related to the CMV isolated from the banana crop in Ecuador (KU127245) with 98.5% identity, followed by the CMV isolated from lulo (Solatium quitoense) in Colombia (MG696856) with 98.3% identity (Gallo et al, 2018). However, it is important to highlight that our CMV quasispecies have a lower identity percentage (between 93.5 and 94.3%) with CMV isolated from tomato-JX101617 in Palmira, Colombia (Uribe-Echeverry, 2012), taking into account that this isolate was found in a plant from the same botanical family and that it was located in the same biogeographic region of the samples evaluated in this study.
TABLE 1 Percentage identities of the 549 bp fragment from CP gene of CMV chili pepper regarding the most related CMV variants found in the Gen-Bank database. The highest identity values are shown in bold.
Considering the classification of the different CMV isolates explained above, CMV sequences of subgroups IA, IB and II were included for our bioinformatics analysis. All reference sequences were downloaded from GenBank. Multiple nucleotide alignment among the chosen sequences and the CMV sequences obtained in this study showed their highest nucleotide identity with a CMV isolated from bananas in Ecuador (KU127245.1) that is placed into subgroup IA. The phylogenetic tree (Fig. 3) confirms that the CMV quasispecies detected in this study belongs to subgroup IA and has a close phylogenetic relationship with the CMV isolate found in banana plants in Ecuador and Colombia. This result has a huge impact because banana was previously grown in some of the areas where our CMV quasispecies was detected. Therefore, it is very possible that the CMV isolate has jumped from banana plants to chili peppers and has successfully adapted to this crop through time and through different selective pressures that operate in chili peppers in relation to those that operate in banana. It is also noteworthy that, although a CMV quasispecies isolate from tomato had been previously detected in Colombia, it is not related to our isolates because the CMV isolated from tomato is clustered in subgroup IB, characterized by having "Asian strains" (Palukaitis and Zaitlin, 1997).
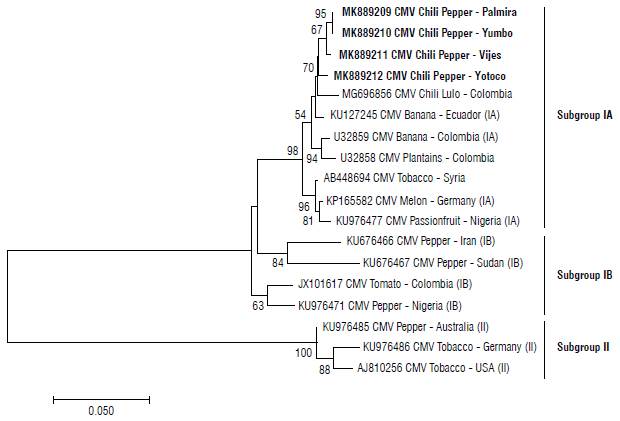
FIGURE 3 Phylogenetic relationships of the CMV chili pepper, constructed from the CP gene at the nucleotide level and its relationship with related CMV isolates. The bar below the tree indicates nucleotide substitutions per site. The evolutionary history was inferred using the neighbor-joining method and the evolutionary distances were computed using the maximum composite likelihood method. The bootstrap consensus of the tree was inferred from 2000 replicates. Only bootstrap values above 50% are shown.
Despite being solanaceous, these results may indicate that the evolutionary selective pressures on the quasispecies found in these crops are different and are leading to speciation that has taken the viral variants so far that they are practically associated with different groups. We cannot discard the possibility that this event could probably happen through the exchange of contaminated seeds, due to the fact that CMV is also disseminated by seeds (Ali and Kobayashi, 2010). To our knowledge, this is the first record of the molecular identification of a CMV quasispecies affecting chili peppers in Colombia.
Specific primer design and detection of CMV chili pepper
With the previously obtained sequences, a pair of primers was designed in order to perform a more precise and sensitive detection of this CMV quasispecies: CMV chili pepper-F (CTT-TAC-GAA-CTG-TCA-CCC) and CMV chili pepper-R (AAC-TAT-TAA-CCA-CCC-AAC-C) that amplified a 229 bp fragment from the CP gene. In order to corroborate the efficacy of these primers, RT-PCR was carried out by taking cDNA from chili pepper samples positive for CMV as well as genomic chili pepper DNA. According to the results obtained in the efficacy test (data not shown), the expected fragment (229 bp) of CMV was observed in the sample that had previously been reported as positive for CMV using the primers described by Herrera-Vasquez et al. (2009). It was also observed that there was no amplification when genomic DNA was used as a template, so the possibility that the amplified fragment corresponds to a part of the genome of the plant is discarded.
With this pair of primers, a new detection on the same samples was carried out. We found that 64.7% (46/71) of the samples were positive for this CMV quasispecies: 100% (6/6) of the samples from Palmira, 62.5% (15/24) of the samples from Yumbo, 73.3% (22/30) of the samples from Vijes and 27.2% (3/11) of the samples from Yotoco. Besides reaffirming the results obtained with the previously described primers, new samples were detected as positive for this virus (Tab. 2). This result shows that the primers designed in this study have high sensitivity, since the number of positive samples for CMV was increased by 12% (64.7%).
TABLE 2 PCR comparison in the CMV chili pepper detection in chili pepper crops using primers reported by Herrera-Vasquez (2009) and primers designed in this work (CMV chili pepper).
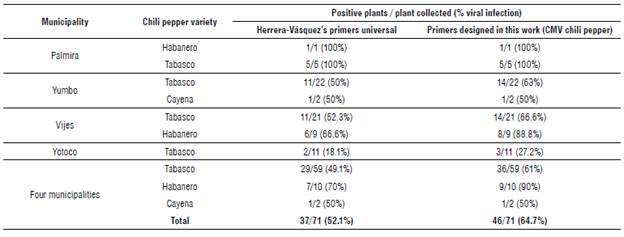
The presence of CMV chili pepper was found in all (three) of the evaluated chili pepper varieties. However, we found that the varieties tabasco and habanero show symptoms associated with viral disease, which could indicate that they are susceptible to CMV chili pepper. Otherwise, the variety cayenne was phenotypically asymptomatic despite being positive for CMV chili pepper. From this result, we can suggest that this variety would serve as a virus reservoir, but it could also be a good material to look for CMV tolerance genes.
Capsid protein analysis of CMV chili pepper
Studies by Perry et al. (1998) have shown that in the capsid protein of CMV there are certain key amino acid positions for viral transmission mediated by vector insects (aphids): one isolate is highly transmissible by aphids (CMV-Fny), and the other one is transmitted at low frequency by aphids (CMV-M), and the amino acids and their position have been determined (Fig. 4). In this study, our CMV isolates were compared and we found that CMV chili peppers shared the amino acid arrangement with the CMV-Fny isolate (Fig. 4) indicating that our isolates most likely belong to the group of those that are highly transmissible by aphids. Also, the study by Perry et al. (1998) shows that some mutations in the conserved positions of the CMV-Fny isolate increase or decrease the transmission by different aphid species, such as Aphis gossypii or Myzus persicae. Our analysis indicates that the CMV chili pepper, in addition to being an isolate highly transmissible by aphids, is more likely to be transmitted by A. gossypii and has a low probability of being transmitted by M. persicae.
Mechanical transmission of CMV chili pepper
At one-month post-inoculation, CMV chili pepper was detected in 86.6% (13/15) of tested plants by RT-PCR, using the specific primers that we previously designed (CMV chili pepper-F / CMV chili pepper-R). These plants showed symptoms such as leaf deformation and yellow vein (Fig. 5). This result confirms that the CMV chili pepper is an infectious agent for chili pepper plants in Colombia.
Conclusions
This study reports, for the first time, the detection and characterization of cucumber mosaic virus in chili pepper crops in four municipalities of Valle del Cauca using molecular techniques in 64.7% (46/71) of the evaluated samples. The CMV chili pepper CP analysis indicated that the virus could be transmissible by the species Aphis gossypii. Also, the CMV primers reported in this study can be implemented as an effective tool in the detection of CMV due to their sensitivity. Finally, this CMV isolate can be considered an infectious agent according to the observed symptoms in the mechanical transmission assay.