Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Iatreia
Print version ISSN 0121-0793
Iatreia vol.22 no.4 Medellín Oct./Dec. 2009
INVESTIGACIÓN ORIGINAL
Asociación del gen TPO en familias colombianas con la diabetes tipo 1
Association of the TPO gene in Colombian families with type 1 diabetes
Javier Gutiérrez–Achury1, Vital Balthazar–González1, Gabriel Bedoya–Berrío2, Andres Ruíz–Linares2,3, Federico Uribe–Londoño4, Juan Manuel Alfaro1, Nicolas Pineda–Trujillo1
1Mapeo Genético, Departamento de Pediatría, Facultad de Medicina, Universidad de Antioquia, Medellin, Colombia. Correspondencia:nicolas.pineda@yahoo.com
2Laboratorio de Genética Molecular (GENMOL), Universidad de Antioquia, Medellín, Colombia.
3Department of Biology, University College London, London UK.
4Departamento de Endocrinología y Metabolismo, Universidad de Antioquia, Medellín, Colombia.
SUMMARY
We have found linkage and association of type 1 diabetes (T1D) to 2p25. The TPO gene lies within this region. Our aim was to test the association of this gene with the susceptibility to T1D in a group of Colombian families, all of them originated in Antioquia, a special population in northwestern Colombia. One hundred familial trios with type 1 diabetes (T1D) were analyzed. They had already been studied for anti–glutamic acid descarboxilase (GAD) antibodies and the marker locus D2S319. For further characterization, the probands were tested for autoantibodies against insulin, TPO and thyrosine phosphatase 2 (IA–2). Two single nucleotide polymorphisms (SNPs) (rs4927611 and rs732609) were tested in TPO. These two markers were chosen considering that the polymorphism changes the encoded amino–acid and a minor allele frequency, MAF, = 0.3. SNP typing was carried out by means of the polymerase chain reaction/restriction fragment length polymorphisms (PCR–RFLP) and the tetraprimer amplification refractory mutation system (ARMS–PCR) methods. Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) analyses were separately tested on both parents and probands. Genetic association was tested by the transmission disequilibrium test (TDT).
It was found that 86% of the patients presented at least one of the three auto–antibodies tested. Both parents and probands were found in HWE for both SNPs. In addition, the two markers were found in tight LD (p < 0.0001). A haplotype associated with susceptibility to the disease was identified (p = 0.0116).
Autoimmunity was found in a proportion similar to that previously reported. It was found that a haplotype at TPO gene is associated with the disease. Such haplotype is characterized by alleles G and A at rs4927611 and rs732609 SNPs, respectively. Our results suggest a possible causative participation of the TPO gene in T1D. In order to verify them, a sample of equivalent size is currently being collected, from the same population in Colombia.
Key words
Autoimmunity, Genetic association, Linkage disequilibrium, TPO gene, Type 1 diabetes mellitus
RESUMEN
Asociación del gen TPO en familias colombianas con la diabetes tipo 1
Hemos encontrado ligamiento y asociación de la diabetes mellitus tipo 1 (T1D) con 2p25. En esta región se halla el gen TPO. Nuestro objetivo fue estudiar la asociación de dicho gen con la susceptibilidad a la T1D en un grupo de familias colombianas, todas ellas originarias de Antioquia, una población especial en el noroccidente del país. Se analizaron cien tríos familiares con T1D. Estas familias ya habían sido estudiadas para anticuerpos contra la ácido–glutámico–descarboxilasa (GAD) y el locus marcador D2S319. Para una caracterización adicional se estudió a los probandos para autoanticuerpos contra insulina, tiroperoxidasa (TPO) y fosfatasa de tirosina 2 (IA2). Se estudiaron en TPO dos polimorfismos de un solo nucleótido (SNP): rs4927611 y rs732609. Se escogieron estos marcadores teniendo en cuenta que el polimorfismo cambia el aminoácido codificado y una frecuencia del alelo menor, MFA, = 0,3. La tipificación de los SNP se llevó a cabo mediante la reacción en cadena de la polimerasa–restricción de fragmentos de longitud polimórfica (PCR–RFLP) y el tetraprimer amplification refractory modification system (ARMS–PCR). Los análisis de equilibrio de Hardy–Weinberg (HWE) y de desequilibrio de ligamiento (LD) se hicieron separadamente tanto en los padres como en los probandos.
Se estudió la asociación genética por medio de la prueba de desequilibrio de la transmisión (TDT). Encontramos que 86% de los pacientes presentaban al menos uno de los tres autoanticuerpos estudiados. Tanto los probandos como sus padres se encontraron en equilibrio de Hardy– Weinberg para ambos SNP. Además, los dos marcadores se encontraron en estrecho desequilibrio de ligamiento (LD) (p < 0,0001). Se identificó un haplotipo asociado con susceptibilidad a la enfermedad (p =0,0116).
Se halló autoinmunidad en una proporción similar a la previamente informada. Se encontró que un haplotipo en el gen TPO está asociado con la enfermedad. Tal haplotipo se caracteriza por los alelos G y A en los SNP rs4927611 y rs732609, respectivamente. Nuestros resultados sugieren una posible participación causal del gen TPO en la T1D. Para verificarlos, se está recolectando una muestra de similar tamaño en la misma población colombiana.
Palabras clave
Asociación genética, Autoinmunidad, Desequilibrio de ligamiento, Diabetes mellitus tipo 1, Gen TPO
INTRODUCTION
Type 1 diabetes (T1D) is a heterogeneous disease; according to the presence of particular auto–antibodies it is classified into autoimmune (T1AD) and idiopathic (T1BD). T1AD is defined by the autoimmune selective destruction of the pancreatic ß cells.1,2 T1BD has no clear cause for the ß cells destruction. T1D is characterized by either a marked or absolute deficiency of insulin, the hormone produced by the pancreatic ß cells,3 which determines the need to replace it with exogenous insulin.
The etiology of T1D is heterogeneous involving both genetic and environmental factors.4 Among the former, HLA is a major susceptibility locus5 and it is accepted that several other non–HLA loci also influence the susceptibility/resistance to T1D.3,6,7 The presence of autoantibodies (Abs) in a risk–HLA allele carrier is associated with a much higher risk for developing the disease.8,10 More often Abs have been described against glutamic acid–descarboxilase (anti–GAD), insulin, islet cells and thyrosine phosphatase 2 (IA–2) antigens.11 In addition, it has been observed that an important proportion of T1D might present anti–thyroid peroxidase auto–antibodies (TPO Abs).12 More recently, the presence of TPO Abs has been associated with specific genetic susceptibility.13
The pathology more often seen simultaneously with T1D is the autoimmune thyroid disease (AITD), which is present in 14% of T1D patients.14,15
One major molecule involved in the AITD onset is the TPO protein (thyroid peroxidase).16 The TPO gene locates at 2p25 and is composed of 17 exons. TPO is a transmembrane protein, of approximately 107 kDa, localized on the apical surface of thyrocytes.16
We have recently found suggestive linkage of T1D to 2p2517 in a large two–generation pedigree with T1D. We found a Lod score of 2.31 at ?=0 at marker locus D2S319. Further genetic analyses allowed to narrow down the candidate region to 3.2 cM. We also tested the marker locus D2S319 in 100 familial trios with T1D from the same Colombian region. It was found that the allele 5 of this marker was associated with T1D (unpublished data). Looking for functional candidates within this region, TPO seems to be a likely gene associated with T1D. In this paper we report further autoimmunity analyses and the evaluation of TPO polymorphisms in a set of one hundred Colombian familial trios with T1D.
PATIENTS AND METHODS
Patients
Our sample was collected from the population living in Antioquia (northwestern Colombia), described as a hybrid population18,19 in which several founder effects have been reported.20,22 The study sample was made up by 100 nuclear families recruited from the index cases (96 families were composed by one affected child and his/her parents. The remainder four families were made up of two–affected sibs and their parents). Patients were identified in the Pediatric Endocrinology Program (Universidad de Antioquia and Hospital San Vicente de Paúl, Medellín, Antioquia, Colombia).
Inclusion criteria were as follows: 1) At least six grandgrandparents born in Antioquia. 2) Both parents of the T1D child were willing to participate in the study. All probands and their parents signed an informed consent approved by the Ethics Committee (Facultad de Medicina, Universidad de Antioquia).
Auto–antibodies evaluation
Auto–antibodies evaluation was carried out in sera samples from the probands. We had previously tested for anti GAD Abs in this sample. Now, we evaluated for Abs against insulin and thyrosine phosphatase 2 (IA–2) by means of an immunofluorescence kit; anti–TPO Abs were tested using a radioimmunoassay kit. In both cases indications of the manufacturer (Kronus) were followed. Considering that our patients had long received treatment with exogenous insulin, the results of this particular Ab were not taken into account.
TPO marker selection and typing
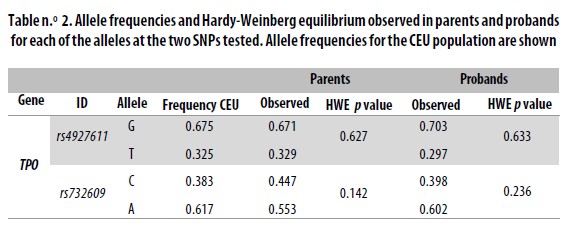
The single nucleotide polymorphisms (SNPs) analized in the TPO gene were chosen from the HapMap database (www.hapmap.org). Selection criteria were: 1) Nonsynonymous SNP, and 2) minor allele frequency (MAF) = 0.3. We selected the SNPs rs4927611 and rs732609. The SNP rs4927611 G<T is located in exon 7 and causes a change of Ser<Ala at position 257 of the protein. The frequencies of its alleles for the HapMap reference population CEU are 0.675 for allele G and 0.325 for allele T. The SNP rs732609 C<A is placed in exon 12 and causes a change of Pro to Thr at position 725 of the protein. Allele frequencies for CEU are A = 0.617 and C = 0.383. Typing of rs732609 was done by PCR/RFLP (polymerase chain reaction /restriction fragment length polymorphisms). The primer sequences used were rs732609–F: CGGGTCATCTG TGACAACAC and rs732609–R: AGCTCCTGGGGAA GATAAGC. The primers were generated using Primer3 (http://frodo.wi.mit.edu/). The PCR fragment was digested with the restriction enzime BsaJI (New England BioLabs) which recognizes the G allele. Digestion products were resolved in agarore gels stained with ethidium bromide.
Typing of rs4927611 was performed with the technique tetraprimer amplification refractory mutation system (ARMS–PCR) described by Ye et al.23 Primers for this technique were generated with the application found at http://cedar.genetics.soton.ac.uk/public_html/primer1.html. Primer sequences will be provided upon request.
Data analysis
Allelic frequencies and Hardy–Weinberg equilibrium
Both markers were analyzed for allele frequencies and Hardy–Weinberg equilibrium (HWE). These analyses were carried out with GENEPOP on the web (http://genepop.curtin.edu.au/)24 using separately the parents and the probands. A p value = 0.05 was considered significant.
TDT analysis
The transmission disequilibrium test (TDT) was performed with the software GENEHUNTER V2.1.25 A p value = 0.05 was considered significant. Mendelian inconsistencies were analyzed with PEDCHECK V1.0.26
RESULTS
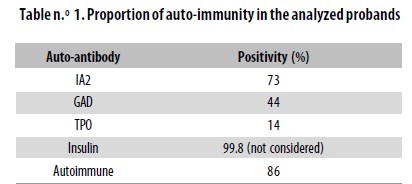
With the information provided by the evaluation of anti– GAD, anti–IA2 and anti–TPO Abs, it was possible to determine that 86% of the sample corresponded to autoimmune T1D (Table n.° 1). A positive result for Abs against TPO was found in 10 individuals (14%). Some sera samples were not tested for this particular Ab.
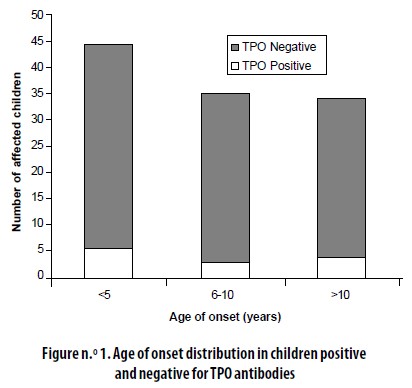
The age of onset distribution among the 10 patients (14%) who were positive for TPO Abs was not different from the one found in the total sample (Figure n.° 1).
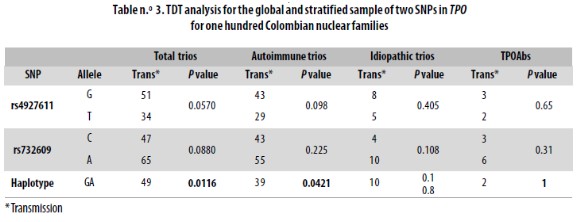
Typing SNPs led to identify marker allele frequencies not significantly different from those reported for CEU in the HapMap database (Table n.° 2). Both markers were in HWE in both the parents and children sets. When analyzed locus by locus in the total sample or when stratified, the transmission disequilibrium test (TDT) was proven not to be significant (Table n.° 3). However, for the haplotype analysis a p = 0.0116 was found in the global sample.
Ver (Tabla1)
Ver (Figura1)
Ver (Tabla2)
Ver (Tabla3)
DISCUSSION
The purpose of this study was to test for the participation of a positional–functional candidate gene at 2p25, associated with T1D susceptibility. This chromosomal region has recently been identified in a meta–analysis of genome wide association studies in T1D.27 Looking at candidate genes in the region of 3.2 Mb mapped by us, TPO seems a plausible gene involved in T1D. Part of its plausibility could be inferred from the observation that a particular genetic variant at CTLA4 is associated with T1D28 and that another variant is associated with autoimmune thyroid disease (AITD).29,30 In like manner, we hypothesized that particular genetic variants at TPO could be associated with congenital hypothyroidism31 and that other variants could explain certain degree of susceptibility to T1D.
An important characteristic of T1D is the presence of autoantibodies. In previous reports its frequency has been between 80–90%.32,33 86% of the probands in our sample were positive for at least one of the Abs tested. When only anti–GAD Abs were considered, only 44% of the sample revealed as autoimmune. However, by testing two more Abs related to the onset of T1D, the proportion of autoimmunity increased approximately two–fold. As a higher positivity rate could be found, it would be interesting to test for other Abs associated with T1D.
Autoimmunity results against insulin were not taken into account because measurements were done after treatment with exogenous insulin had been started. Consequently, Abs against insulin could be due to either an autoimmune process or to a reaction against exogenous insulin.
Fourteen percent of our T1D patients were positive for TPO Abs. This finding is close to that reported by other authors.12 However, we have no certainty on whether TPO Abs had appeared at onset of T1D or later.
Our TDT analysis did not reveal any significant association when we analyzed a marker locus at a time. However, by analyzing the haplotypes it was suggested that alleles G and A at rs4927611 and rs732609, respectively, were transmitted together to affected children more often than expected by chance. In the global sample, after correction for multiple tests (0.05/4), the significance value is just kept to the 95% confidence (p < 0.0116). In the stratified sample, p value was higher (p = 0.0421) than when taken together (total trios p = 0.0116); this could be due to the decrease in the sample size.
Even though the signal is weak, it can be evidence that the TPO gene is involved in the susceptibility to T1D in Antioquia. A larger sample size might help define this association. In addition, replication in an independent sample would confirm this finding. Another possible scenario includes other gene from the candidate region identified in 2p25, which could be in linkage disequilbrium with TPO.
AKNOWLEDGMENTS
This study was supported by Colciencias, grant n.° 111534319156 and CODI – Universidad de Antioquia, grant n.° 8704–2449. We are very grateful to Doctor Juan Manuel Anaya for having kindly typed the autoantibodies against TPO for this study.
BIBLIOGRAPHIC REFERENCES
1. Jansson SP, Andersson DK, Svardsudd K. Prevalence and incidence rate of diabetes mellitus in a Swedish community during 30 years of follow–up. Diabetologia 2007; 50: 703–710. [ Links ]
2. Park Y. Functional evaluation of the type 1 diabetes (T1D) susceptibility candidate genes. Diabetes Res Clin Pract 2007; 77 (Suppl. 1): S110–115. [ Links ]
3. Alizadeh BZ, Koeleman BPC. Genetic polimorphism in susceptibility to type 1 diabetes. Clin Chim Acta 2008; 387: 9–17. [ Links ]
4. Todd JA. Genetic control of autoimmunity in type 1 diabetes. Immunol Today 1990; 11: 122–129. [ Links ]
5. Steenkiste A, Valdes AM, Feolo M, Hoffman D, Concannon P, Noble J, et al. 14th International HLA and Immunogenetics Workshop: report on the HLA component of type 1 diabetes. Tissue Antigens 2007; 69 (Suppl. 1): 214–225. [ Links ]
6. Bruno G, Biggeri A, Merletti F, Pagano G. Temporal trends in incidence rates of type I diabetes in Germany: birth cohort and calendar period effects. Diabetologia 2000; 43: 1334–1336. [ Links ]
7. Ounissi–Benkalha H, Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med 2008; 14: 268–275. [ Links ]
8. Bugawan TL, Klitz W, Alejandrino M, Ching J, Panelo A, Solfelix CM, et al. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens 2002; 59: 452–469. [ Links ]
9. Cucca F, Lampis R, Congia M, Angius E, Nutland S, Bain SC, et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet 2001; 10: 2025–2037. [ Links ]
10. Thomson G, Valdes AM, Noble JA, Kockum I, Grote MN, Najman J, et al. Relative predispositional effects of HLA class II DRB1–DQB1 haplotypes and genotypes on type 1 diabetes: a meta–analysis. Tissue Antigens 2007; 70: 110–127. [ Links ]
11. Pietropaolo M, Barinas–Mitchell E, Kuller LH. The heterogeneity of diabetes: unraveling a dispute: is systemic inflammation related to islet autoimmunity? Diabetes 2007; 56: 1189–1197. [ Links ]
12. Gonzalez GC, Capel I, Rodriguez–Espinosa J, Mauricio D, de Leiva A, Perez A. Thyroid autoimmunity at onset of type 1 diabetes as a predictor of thyroid dysfunction. Diabetes Care 2007; 30: 1611–1612. [ Links ]
13. Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, Todd JA. A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T–lymphocyte–associated antigen–4 gene. Diabetologia 2007; 50: 741–746. [ Links ]
14. Barker JM, Yu J, Yu L, Wang J, Miao D, Bao F, et al. Autoantibody 'subspecificity' in type 1 diabetes: risk for organ–specific autoimmunity clusters in distinct groups. Diabetes Care 2005; 28: 850–855. [ Links ]
15. Kordonouri O, Hartmann R, Deiss D, Wilms M, Gruters– Kieslich A. Natural course of autoimmune thyroiditis in type 1 diabetes: association with gender, age, diabetes duration, and puberty. Arch Dis Child 2005; 90: 411–414. [ Links ]
16. McLachlan SM, Rapoport B. Thyroid peroxidase as an autoantigen. Thyroid 2007; 17: 939–948. [ Links ]
17. Uribe F, Pineda–Trujillo N, Montoya F, Latorre G, Villegas A, Ceron J, et al. Análisis de ligamiento genético de la diabetes mellitus tipo 1 a marcadores de los cromosomas 2 y 11 en familias antioqueñas. Iatreia 2004; 17: 93–104. [ Links ]
18. Carvajal–Carmona LG, Ophoff R, Service S, Hartiala J, Molina J, Leon P, et al. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet 2003; 112: 534–541. [ Links ]
19. Carvajal–Carmona LG, Soto ID, Pineda N, Ortiz– Barrientos D, Duque C, Ospina–Duque J, et al. Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am J Hum Genet 2000; 67: 1287–1295. [ Links ]
20. Lopera F, Ardila A, Martinez A, Madrigal L, Arango–Viana JC, Lemere CA, et al. Clinical features of early–onset Alzheimer disease in a large kindred with an E280A presenilin–1 mutation. Jama 1997; 277: 793–799. [ Links ]
21. Pineda–Trujillo N, Apergi M, Moreno S, Arias W, Lesage S, Franco A, et al. A genetic cluster of early onset Parkinson's disease in a Colombian population. Am J Med Genet B Neuropsychiatr Genet 2006; 141: 885–889. [ Links ]
22. Pineda–Trujillo N, Carvajal–Carmona LG, Buritica O, Moreno S, Uribe C, Pineda D, et al. A novel Cys212Tyr founder mutation in parkin and allelic heterogeneity of juvenile Parkinsonism in a population from North West Colombia. Neurosci Letters 2001; 298: 87–90. [ Links ]
23. Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 2001; 29: E88. Disponible en http://www.ingentaconnect.com/conten/oup/nar/2001/00000029/00000017/e88 Consultado 12 de noviembre de 2009. [ Links ]
24. Raymond M, Rousset F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J Hered 1995; 86: 248–249. [ Links ]
25. Kruglyak L, Daly MJ, Reeve–Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996; 58: 1347– 1363. [ Links ]
26. O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998; 63: 259–266. [ Links ]
27. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome–wide association study and meta–analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41: 703–707. [ Links ]
28. Nistico L, Cascino I, Buzzetti R, Pozzilli P. CTLA–4 in type 1 diabetes. En: Madame Curie Bioscience Database, disponible en http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=eurekah&part=A15486. Consultado el 20 de agosto de 2009 [ Links ]
29. Kouki T, Sawai Y, Gardine CA, Fisfalen ME, Alegre ML, DeGroot LJ. CTLA–4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA–4 and contributes to the pathogenesis of Graves' disease. J Immunol 2000; 165: 6606–6611. [ Links ]
30. Yanagawa T, Hidaka Y, Guimaraes V, Soliman M, DeGroot LJ. CTLA–4 gene polymorphism associated with Graves' disease in a Caucasian population. J Clin Endocrinol Metab 1995; 80: 41–45. [ Links ]
31. Tenenbaum–Rakover Y, Mamanasiri S, Ris–Stalpers C, German A, Sack J, Allon–Shalev S, et al. Clinical and genetic characteristics of congenital hypothyroidism due to mutations in the thyroid peroxidase (TPO) gene in Israelis. Clin Endocrinol 2007; 66: 695–702. [ Links ]
32. Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2000;23 (Suppl. 1): S4–19. [ Links ]
33. Narendran P, Estella E, Fourlanos S. Immunology of type 1 diabetes. QJM 2005; 98: 547–556. [ Links ]
Recibido: julio 09 de 2009
Aceptado: octubre 16 de 2009