I. INTRODUCTION
Agriculture is one of the most important economic industries for the internal production of a country since it makes possible the production of food and various products that depend primarily on small farmers, who are responsible for 80% of the general production. Unfortunately, at least 50% of the yield of this production is lost due to crop pests and diseases [1]. However, automated systems capable of identifying the disease type and providing timely support to take appropriate action have been developed to address these issues and benefit agriculture [2].
In recent years, different fast and effective methods have been proposed to detect diseases in plants and thus avoid losses in the agricultural industry [3]; for example, by the end of the '70s, computer image processing technology was applied to agricultural studies. Researchers demonstrated that these techniques could be used as a disease detection system [4]. Later on, between the '80s and '90s, remote detection and technical diagnosis active applications based on image processing were developed to create detection algorithms with machine learning [5]. Thus, it is evident the growing interest in researching the application of machine learning algorithms to identify plant diseases. Several machine learning techniques have been used and discussed; nevertheless, it seems there is not a universal technique for all types of situations [6].
The world population is expected to increase to 9 billion by 2050; therefore, developing methods for detecting and mitigating plant diseases has two purposes that favor food security: increasing the crops yield and reducing the use of pesticides [7]. While the accuracy of traditional detection methods, such as the PCR, is not disputed, a system that uses one of these artificial intelligence structures can offer an early detection method to be used along with traditional laboratory tests, expediting responses and mitigating crop losses [8]. Furthermore, researching the detection of plant diseases benefits farmers since many of them worldwide do not have access to technical advice for rural areas, making their crops especially vulnerable to yield losses and other problems caused by plant diseases [9].
On the other hand, regarding theoretical research and practical applications, image classification for disease detection through crop leaves is of great importance [10] and, therefore, it is the focus of this study. Many researchers have studied crop disease detection based on pattern recognition and found that the farmers’ knowledge can be supported by data engineering and tools or techniques associated with several agriculture sectors to improve their productivity [11].
This paper aims at providing a vision of the different techniques used for plant disease detection from an accuracy and interpretability perspective. Additionally, it presents a summary of factors and processes commonly used when researching this topic. Initially, a literature review protocol that allows obtaining a set of articles with a high level of relevance regarding the main topic of this study is outlined. Then, a series of core questions to guide the purpose of this study are proposed. Subsequently, the search of the literature in different databases is carried out considering specific selection criteria. Finally, the scope and impact of the results obtained through this research are discussed. The rest of the document provides a detailed analysis of the different factors related to plant disease detection from an interpretability perspective and answers the initial research questions.
II. METHODOLOGY
To determine the current state of the machine learning techniques used for the detection and classification of plant diseases, it is necessary to define a series of steps to follow. Following Massaro’s structured literature review (SLR) [12], where a guide for literature review is provided, the following steps are proposed:
Literature review protocol
Defining core questions to be answered by the literature review
Searching for literature related to the research subject
Defining the article’s impact
Establishing the analytical framework
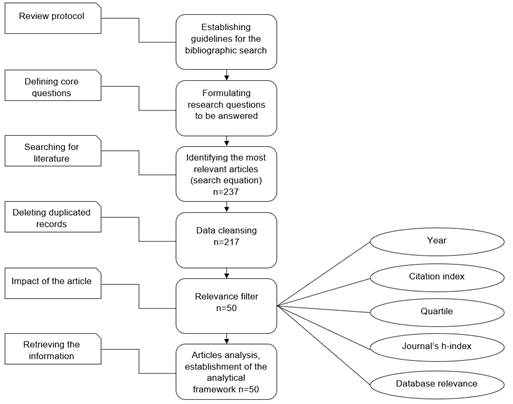
The previous steps can be observed in Figure 1, where the process to obtain a set of adequately filtered literature is described in detail. Then, the analytical framework is established to obtain the results that allow to determine the state of the research in the area and answer the research questions.
A. Bibliographic Review Protocol.
This bibliographic review is focused on the articles most relevant for the main subjects related to image analysis, plant diseases, detection, and machine learning. A total of 51 articles were weighted based on the quartile of their respective journal, the article’s citation index, the journal’s h-index, the article’s year of publication, and their relevance based on the search equation of each database. These parameters will allow to select and reduce the total number of research papers to be analyzed, thus, guiding this literature review toward the most relevant articles in the research field.
B. Research Questions Definition.
The following questions aim at guiding the literature review:
- Which machine learning methods are currently the most widely used for plant disease detection using image analysis techniques?
- What are the trends in machine learning methods used for plant disease detection using image analysis techniques?
- What is the role of interpretability in the machine learning methods developed by the authors?
C. Literature Search.
Once the literature review protocol and the research questions are defined, the search equation is created to obtain the largest number of articles related to the research subject, as shown in Table 1. A total of 226 articles were obtained in the academic databases Scopus, Web of Science, Science Direct, and Google Scholar.
Table 1 Search equation.
| Operator | Keywords | Limited to |
|---|---|---|
| OR | ("plant* diseas*" OR (diseas* AND image AND plant*) OR "plant* leaf" OR "crop* diseas*") OR KEY ("plant* diseas*" OR (diseas* AND image AND plant*) OR "plant* leaf" OR "crop* diseas*")) | Tittle |
| ("plant* diseas*" OR (diseas* AND image AND plant*) OR "plant* leaf" OR "crop* diseas*") OR KEY ("plant* diseas*" OR (diseas* AND image AND plant*) OR "plant* leaf" OR "crop* diseas*")) | Keyword | |
| AND | ||
| OR | ("image analysis" OR "computer vision" OR "Image classification" OR " Image segmentation" OR "Image Processing") | Tittle |
| ("image analysis" OR "computer vision" OR "Image classification" OR " Image segmentation" OR "Image Processing") | Keyword | |
| AND | "detect*" | Tittle |
1) Initial Cleansing. To delete possible duplicated records, the obtained data were cleansed based on the article’s title and authors, which resulted in 20 duplicated records for a total of 206 articles.
2) Defining the Article’s Impact. For the selection process of the most relevant and significant literature for this research, a weighting of different relevant factors from each article is established, among which are: year, citation index, quartile, h-index for the journal, and the relevance of each article given by the database from which it was extracted. In Table 2, each factor has its corresponding relevance for this search.
III. ANALYSIS
In this section, different aspects of the 51 articles selected in the bibliographic review are assessed.
A. Disease Detection in Plants.
This section presents a perspective of the systems developed for disease detection that reduce the follow-up work in extensive crop areas, which in early stages is beneficial if the symptoms are detected when they appear in the leaves [13]. However, although in different articles the detection and classification were considered altogether, it is evident that for several researchers, the most important one is detection, since it is the main objective of the methods developed by computer systems, as shown in the analysis of each of the stages implemented to detect diseases described below.
B. Data Preparation.
1) Image Acquisition. To detect diseases under any circumstances, authors acquired images in an adequate or random environment to train and test a model. Table 3 summarizes the sources used in the acquisition of images and a numerical range to indicate how many images were obtained by each source in the reviewed articles.
Table 3 Summary of the image acquisition sources.
| Guide references | Image acquisition source | Acquired images range |
|---|---|---|
| [14], [15] | Unmanned aerial vehicle (UAV) | 340 - 17640 |
| [3], [9], [11], [13], [16]-[19] | Digital cameras | 106 - 9000 |
| [2], [8], [10], [20]-[24] [22], [25]-[27] | Plant Village and websites | 733 - 54305 |
| [8], [22], [25]-[33] | Others (laboratory images, satellite images, hyperspectral images) | 180 - 33469 |
These sources usually belong to open-access databases to enable, in combination with advances in computer vision, the deciphering of specific symptoms of plant diseases [2]. It is noticeable that most of the images acquired by the researchers are focused on crops that are more susceptible to diseases that mainly affect their leaves.
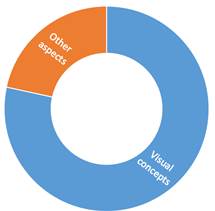
2) Preprocessing. In this step, researchers use several techniques that highlight the affected regions based on elements such as temperature, noise, contrast, and others, trying to make the image more visible and easier to understand compared to the original. Figure 2 shows the aspects considered to make the images suitable for entering the detection model and thus improve its accuracy when determining if a plant is sick.
Forty of the papers analyzed improved the quality of the image based on visual concepts. Usually, the initial images are in an RGB color model, and the transformation depends on how the author considers the model will have better accuracy. Table 4 presents the different preprocessing techniques reviewed, emphasizing the techniques found according to the visual concept processing.
Table 4 Summary of the image preprocessing techniques of the papers reviewed.
| Technique | Model | Accuracy interval | Number of articles |
|---|---|---|---|
| Color transformation | CIELAB | 58 - 99.7 | 9 |
| Infrared color | > 80 | 1 | |
| HSV | 58 - 99.7 | 4 | |
| H, I3a and I3b | > 83.5 | 1 | |
| Monochromatic | 93.9 - 98.51 | 3 | |
| YCbCr | 88.9 - 98.51 | 4 | |
| HSI | 88 - 97.6 | 5 | |
| CIELUV | > 95.8 | 1 | |
| CMYK | 58 - 90 | 2 | |
| Image enhancement | Hyperspectral images | > 98 | 1 |
| Multispectral thermal images | 92.81 - 93.55 | 2 | |
| Enhanced RGB | 60 - 99.7 | 32 |
One image preprocessing example is presented in [26], where the Ycb, HSI, and CIELAB color models were used to detect disease spots successfully without being affected by the noise. Similarly, in [16] and [22], some techniques that transform color to CIELAB, HSV, and HSI models are used to perceive and interpret better the images helping the model detect explicit information more efficiently by eliminating lighting and contrast issues.
3) Segmentation. Image segmentation is one of the most critical stages of the pattern recognition process since it affects the classification results [24]. In this stage, the image is divided into regions with particular relevance to train the model with the help of different techniques based on artificial vision, which assigns a special meaning to the fragments of the image, then, a category is assigned to each pixel as the model analyzes an image.
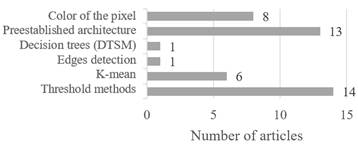
Forty-three of the 51 papers reviewed specified the technique for the segmentation of the plants’ images and the automatic detection of lesions in them, which resulted in data labelling, pattern identification or similarities as appropriate. Figure 3 shows the techniques most commonly used by researchers to develop this stage during the data preparation, considering the relevance of each of them in the machine learning development within each computerized system. There are many different ways of carrying out image segmentation; these range from simple thresholding to advanced color image segmentation methods.
C. Feature Selection.
1) Feature Extraction. In this section, the main problem addressed is finding an accurate representation of the image of a plant affected by disease using feature extractors [24]. This stage is one of the most complex since the noticeable features such as texture, shape, size, and color are recognized in all the images used to train a model. Furthermore, obtaining good results in the validation and testing with different images depends on this. Therefore, feature extraction plays an essential role in artificial vision and pattern recognition to represent an image [22].
To fulfil the purpose of reducing and specifying the necessary data, the researchers consulted have established or designed diverse architectures based on the features of the lesions in plants, looking for the best performance, as shown in Table 5.
Table 5 Papers reviewed with the main identified architectures used in machine learning.
| Architecture | Performance interval | References |
|---|---|---|
| Hybrid-type intelligent algorithm | 92 - 99.7 | [16], [17], [19], [21], [22], [25], [35], [36] |
| ResNet and U-Net deep learning architecture | 93 - 98.51 | [8], [21], [37] |
| Visual geometry group | 87.2 - 96 | [20], [38] |
| AlexNet | 97.39 - 98.51 | [1], [8] |
| Specialized models | 92 - 93.72 | [20], [34] |
| Based on specialized generic modules | > 83 | [39] |
| AdaBoost algorithm | > 88 | [18] |
Feature extraction techniques have been a significant evolutionary development in areas such as agriculture since they reduce the redundancy of the information and contribute to the execution time, making crop disease detection a faster and more efficient process. Nevertheless, during this review, it was evidenced that for the execution of these techniques, researchers design their algorithms in specific coding platforms whose features contribute to machine learning, among which the following stand out: Matlab, Python, Opencv, C++, and TensorFlow.
2) Learning Technique. Finally, this is the stage related to the learning task. Its main objective is to identify patterns so that the entry data are classified in several groups repetitively based on the default algorithms for the system, continuously improving the process according to the relevance of the data.
The machine learning techniques are in charge of this task, as they have proven to be efficient classifiers in diverse applications throughout their history. Table 6 summarizes the main techniques used by the researchers consulted in this review for the data classification and disease detection, each trained for specific plants and diseases. Likewise, it is essential to consider that some models, such as the one proposed in [1], classify the data with the help of two or more learning techniques to optimize the results, which affects the accuracy percentage and the learning task carried out by these methods.
Table 6 Machine learning techniques in the papers reviewed according to their learning task and accuracy.
| Machine learning technique | Number of articles | Accuracy interval | Guide references |
|---|---|---|---|
| Support vector machine (SVM) | 26 | 54.1% - 99.7% | [3], [17], [42] |
| KNN | 22 | 67.9% - 98.4% | [11], [25], [36], [38], [43] |
| Convolutional neural network | 17 | 84.8% - 98.6% | [15], [23], [41], [44] |
| Random forest | 13 | 54.1% - 99.7% | [1], [14], [29], [45] |
| Neural network | 8 | 60% - 98% | [5], [19], [21], [40] |
| Naïve Bayes | 8 | 60% - 98.4% | [14], [35], [46] |
| Decision tree | 4 | 54.1% - 99.7% | [16], [30], [32], [47] |
| Deep neural network | 3 | 92% - 98.3% | [20], [48] |
| Probabilistic neural network | 2 | 96% - 98.4% | [20], [35] |
| Backpropagation neural network | 2 | 89% - 98.33% | [4], [45] |
| Boosted regression tree | 2 | 87.2% - 95% | [19], [31] |
| Logistic regression | 1 | 95% - 97% | [19] |
D. Machine Learning Approaches.
In general, early disease detection models based on machine learning solve the complex issue of recognizing patterns in images of infected or healthy leaves employing different techniques or models as required. Figure 4 presents the machine learning approach used in the 51 articles reviewed. Some models have been trained and tested with more than one approach to see their behavior and optimize the results. Nonetheless, those were included to observe the authors’ tendency toward these approaches when training their plant disease detection algorithms.
E. Model Optimization.
This section groups the optimization techniques used in the articles reviewed. It becomes evident that the main objective of implementing these techniques is to reduce the losses and provide the most accurate results, hence the importance of considering them during the review. In addition to being aware of how the proposed models work, it is relevant to know that it is common to combine them with other learning functions that help adjust some values or are developed based on a specific algorithm to improve their accuracy. In this case, 33 of the models reviewed were optimized with the techniques described in Table 7, whereas the rest of the papers did not optimize their models since they obtained an acceptable performance without implementing these techniques during the learning task determined by each author.
Tabla 7 Summary of the models’ optimization techniques used in the papers reviewed.
| Optimization technique | Number of articles | Some algorithms used | References |
|---|---|---|---|
| Stochastic gradient descent | 9 | Without variations in the review | [20], [21] |
| Customized optimizers | 9 | Graph cutting algorithm, GLCM algorithm Delta E algorithm and speeded-up robust features (SURF) | [4], [5], [16], [49] |
| Particle swarm optimization | 2 | Particle swarm optimization (PSO) | [1], [3] |
| Gradient descent optimizer | 7 | Adam optimization algorithm | [7], [19], [47] |
| Successive projections algorithm | 3 | Without variations in the review | [31], [45], [50] |
| Function selection algorithm | 1 | Algorithm Relief-F | [51] |
| Segmentation algorithm | 2 | Mean-shift segmentation algorithm | [14], [39] |
| Iterative algorithm | 3 | Expectation maximization algorithm and simple linear iterative clustering | [15], [36], [52] |
| Genetic algorithm | 3 | Without variations in the review | [40], [53] |
F. Performance Assessment Methods.
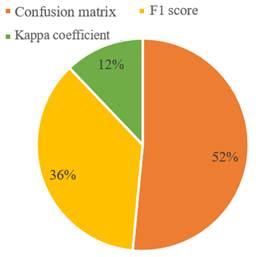
In machine learning, it is necessary to assess the general performance of a proposed model to know its pattern recognition ability and thus determine if it effectively complies with the assigned task. Although a variety of performance measures are used for that purpose, three specific measures were considered for this review, which have proven to be the most relevant and accurate to develop this task when considering the sensitivity to atypical values, the execution time, and even the true and false positives.
Figure 5 shows the usage percentage of the confusion or error matrix, F1 score, and the Kappa coefficient methods, which were outstanding and cross-sectional methods in the review to verify the performance of each method.
Some researchers assess their model by using several review metrics to verify and maintain a follow-up on the data since their objective is to improve the general prediction of each model before working with new data.
G. Interpretability.
Finally, in the literature reviewed, it was found that only one author [35] addressed the issue of interpretability in their proposed model by developing a hybrid system composed of techniques known for their high accuracy and interpretability, such as vector support machines and neural networks, in addition to Mamdani-type diffuse systems. This way, the author addressed the construction of a hybrid model that allows the end-user to know part of the composition of the model and analyze its deductions without neglecting accuracy.
IV. DISCUSSION
All the articles reviewed for this research meet the requirement of being models designed for the early detection of plant diseases in a non-destructive way based on images. These images were acquired from different crops and processed in various ways. However, there is a void in the interpretability field between the accuracy and interpretability of each model. Only 1 out of 51 articles is highly interpretable as it explains in detail its functionality and simplifies the complexity of understanding how the proposed model makes decisions. This void results in users not knowing what affects the results or the features recognized in each model.
During this review, it was also possible to evidence the gradual and significant progress of the image processing techniques applied to images from different crops. These are usually obtained by digital cameras in controlled environments, web databases such as PlantVillage and even hyperspectral images taken by UAVs, and then processed based on their visual concepts and color transformation models such as CIELAB and image enhancement such as enhanced RGB.
In developing machine learning models, image segmentation is vital to differentiate sick regions in plants. The most common techniques used by the researchers for this purpose are based on preestablished architectures and threshold methods, which accomplish the task of separating the relevant objects of an image from the rest when the image is clearly visible. In the same way, after this image segmentation, the algorithm must comply with the task of reducing and specifying the necessary data. The hybrid algorithms and deep learning architectures such as ResNet and U-Net are the most prominent.
In the feature classification stage, where the objectives are accuracy and efficiency, it is possible to answer the research question: Which machine learning methods are currently the most widely used for plant disease detection using image analysis techniques? In table 6, the methods used were organized from most to least used by the researchers, showing that the vector support machines, KNN, and convolutional neural networks methods are the most outstanding due to their accuracy in machine learning to detect diseases and classify plants as healthy or sick.
Similarly, this review answered the question: What are the trends in machine learning methods used for plant disease detection using image analysis techniques? Figure 4 summarizes the machine learning approaches used for plant disease detection through image analysis techniques, considering the traditional approaches and an additional one called transfer learning. This shows there is a trend toward using supervised learning.
Finally, another question was answered: What is the role of interpretability in the machine learning methods developed by the authors? It is concluded that most of the current models and those in development are focused mainly on high accuracy, leaving aside the role of interpretability since if it is not addressed correctly, it can significantly affect the accuracy of the models.
Although there are algorithms with good performance in detecting diseases without additional functions, most of the researchers prefer to optimize their models with the techniques described in Table 7. Two that stand out are the stochastic gradient descent and the gradient descent optimizer, which minimize the data loss, providing more accurate results due to all the training data and use the loss of only one sample from training to calculate the average loss.
V. CONCLUSIONS
When gathering the most critical data in the development of an automated system that detects plant diseases through images, it is concluded that machine learning is an appropriate assistance for farmers since early disease detection allows them to make better decisions for their crops and improve their productivity. Additionally, this type of research significantly increases the studied species’ scope, the techniques developed, and the classified diseases.
Similarly, it is concluded that there is a wide range of alternatives to develop a detection model using pattern recognition since there are various ways of capturing images, focusing on relevant areas in them and developing algorithms, supervised or not that contribute to the accuracy, in this case of agriculture, and thus improve the yield in each research continuously.
This review also allowed to notice a void in the interpretability field since the authors creating disease detection models through images have been focused mainly on obtaining good results, overlooking the possibility of giving the user a clear explanation of the features recognized or the specific issues affecting their results. Therefore, it is important to consider this element in the development of future models since it is relevant for the user to know the structure of the model and be able to deduce from it the process to obtain the results. This can help in the creation of more relevant models that improve agriculture.