I. INTRODUCTION
Biological collections contain important information on Colombian biodiversity and are a national heritage. Through them, knowledge can be generated in the management, conservation and sustainable use of biodiversity. They are a cumulative record of knowledge generated at a specific time and place, allowing us to observe our territory's natural wealth over time [1]. Colombia is a megadiverse country where the description of new species will greatly affect the global knowledge of biodiversity [2]. However, it is facing an accelerated rate of extinction of species, where the loss of species is greater than the knowledge about them [3].
Carrying out a complete inventory of biodiversity implies describing unknown and known species, understanding the diversity of biology and creating databases to handle information systematically. Computer networks are also created for the flow of information. All this is important due to the lack of knowledge about the state of biodiversity in a region [4]. This research project seeks to solve the lack of information management in the UNISANGIL - CBMUS Macroinvertebrate Biological Reference Collection. Instead of addressing all aspects related to the biology of the species, the focus is on the technical aspects of the tool to support macroinvertebrate research.
The application allows knowing the characteristics of macroinvertebrates and recording data for researchers to estimate biochemical, ecological and biotic factors. Solutions such as ecosystem bioremediation can also be implemented. This technological development seeks to create an updatable and reliable database so that researchers, universities, research institutes and decision makers can share investigative processes.
Bioinformatics is dedicated to applying informatics in managing information related to biological and medical data through the collection, storage, organization, analysis, manipulation, presentation and distribution of information. Its objective is to act as a bridge between data and information, allowing to obtain knowledge about biological processes from data analysis [5]. Currently, technology provides important preservation tools that help to save an original in a standard format that does not depend on a special technology for its recovery. This tool offers the opportunity to preserve the original by providing access to the digital substitute and separating the informational content from the degradation of the physical medium [6]. Historical documents are a precious asset for the institutions that protect them, and it is common to seek their preservation and conservation to keep them in good condition. These archives are of great value and often unavailable to the general public [7].
Digitization consists of converting an image to binary code with the help of a computer, providing a detailed description of the objects in the scene. Each pixel is assigned a tonal value represented by a binary code, and these binary digits are stored and often reduced to a mathematical representation. The computer then interprets the bit stream to reproduce an analog version that can be displayed or printed [8]. Biological collections are an important store of information on biodiversity. The extended specimen preserves the secrets of the natural world and the memory of the environment.
Given the global difficulties in the conservation of species, collections are becoming increasingly important since biological records and specimen information have helped categorise threats to endangered species [9]. The National Registry of Biological Collections (RNC) collects and disseminates basic information on biological collections in the country, which makes its invaluable natural heritage visible. This information is essential for the management of biodiversity and ecosystem services [10].
The Darwin Core Standard (DwC) was developed by the Biodiversity Information Standards community and is used by the Biodiversity Information System in Colombia. This standard allows for the compilation of biodiversity information from different sites and is used by most of the species presence records available on GBIF.org. The related file format is called Darwin Core Archive (DwC-A) and allows data providers to share their information using a common lexicon.
This standardization not only reduces the process of publishing biodiversity information but also facilitates the investigation, evaluation and comparison of data to find solutions [11]. Aquatic macroinvertebrates are large invertebrates measuring more than 500 μm. In freshwater bodies, insects are the most diverse group of aquatic invertebrates, with aquatic immature stages and terrestrial adults. Mayflies, stoneflies and odonates are some of the most common and widely distributed aquatic insects [12].
Software is crucial to carry out various academic, research and industrial operations, and new applications are developed to respond to the needs or opportunities of communities. Software design, development, and implementation processes are essential in business systems [13]. Agile methodologies for software development provide a set of techniques to be applied in stages in short work cycles, with the aim of making the project delivery process more efficient, without having to wait until the end [14]. Agile methods favor incremental development, generating new releases every two or three weeks, which are available to customers to get feedback according to requirements quickly [13].
The agile methodology focuses on continuous improvement through planning, creation, verification and constant improvement, with rapid deliveries to avoid dispersion and focus on an entrusted task [15]. Scrum is an agile software development methodology based on an iterative and incremental approach, made up of sprints that can last between one and four weeks. At the beginning of each sprint, a meeting is held to define the work to be developed, followed by daily reviews to verify the progress in the development of the sprint and at the end of it, another meeting is held to evaluate the results [16].
A Scrum is divided into three phases: planning, sprint cycle development, and project closure. During planning, the objectives of the project and the software architecture are established, the sprint cycles generate the system increments, and in the closing phase, the complete functionality is delivered together with the documentation [13]. For the development of the project, various tools were used, such as PostgreSQL for database management and programming languages such as Javascript, Typescript and CSS for designing user interfaces in a web environment. In the field of biological collections, there are insufficient or expensive applications to protect the information of the specimens.
For example, Specify requires knowledge of databases, programming languages, and markup languages. It is challenging to maintain trained personnel for this purpose; thus, interdisciplinary collaboration is important to develop more economical and efficient solutions that respond to the needs of the collections.
II. METHODS
At UNISANGIL, the framework proposed by Dr. Roberto Hernández Sampieri is used to carry out research processes. The Faculty of Natural Sciences and Engineering follows this approach to define the problem, establish the relationship between variables and formulate the research question. In the same way, the objectives of the project and the methodology were defined, a schedule of activities was designed to monitor the execution of the project and its deliverables were established, which resulted in the development of the bioinformatics platform [17].
Consequently, a question was raised: How to make the UNISANGIL CBMUS aquatic macroinvertebrate biological reference collection known to the community? The agile Scrum methodology was selected for software development, and frequent meetings were held with the client to manage the application requirements and model them using UML diagrams [18]. The phases considered were: requirements management, backlog management, sprint planning and execution, and finally, inspection and iteration [19]. The actions carried out in each phase are detailed below.
A. Definition of the Application Requirements
In this stage, sources of information and applications available to manage biological collections such as Specify, Simbiota and Elysia were explored, and sessions were organized with biologists and researchers from the CBMUS macroinvertebrate collection to define the application requirements. The Darwin Core standard was chosen to design and create the information storage structure based on the advice and support of the Humboldt Institute. Finally, four functionalities were defined for the application: access control, user administration, information registry, and query module. Based on this definition, the requirements for each system module were collected and prioritized in templates. Work was also done on the definition of user stories that allowed better analysis of the system and later identification of each of the actions that users perform on the platform.
B. Backlog Management
In this stage, the list of functionalities and tasks to be carried out was generated, allowing to obtain the details of the actions to be implemented for the creation of each of the identified modules, together with the allocation of time and those responsible. From the definition of the four modules that make up the platform, it was decided to guide the development process for each functionality.
C. Sprint Planning Meeting
During the first iteration, the focus was on planning the project. The highest priority requirements for each module were selected, and the application development phase started. A list of actions necessary for the development of each module was prepared, and work was done on the requirements identified in the previous stage. In addition, the effort estimate was made, and times and responsibilities were assigned to continue with the next phase of the project.
D. Sprint Execution
The Scrum methodology was used, and the project was divided into four sprints. Frequent meetings were held to assess the team's progress and troubleshoot. We worked with the questions: What have I done since the last synchronization meeting? What am I going to do from now on? And what impediments do I have or will I have?
E. Inspection and Iteration
After completing each iteration of the project, the team met with the client to present the fulfilled requirements. The client evaluated and reviewed the delivery and approved the increases or indicated the necessary adjustments. Then, in a retrospective meeting, the team would evaluate their performance and apply what they learned in the next phase. The platform database was designed taking as a reference the Darwin Core standard used by SiB Colombia during the enlistment process of the different components [20]. After some tests, it was decided to use a Dashboard type interface for the project, with the aim of providing a pleasant user experience. The first user interface designs were made using the Balsamiq Wireframes software [21], and tests were carried out to choose the best option.
III. RESULTS
As a result of the development of the project, the following was achieved.
A. Application Architecture
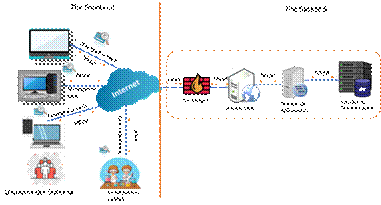
The architecture of the platform was defined to provide comfort, friendliness and security features to the user and to host the information transparently and independently from the administration staff. It was decided to work with a web server client architecture that would allow hosting and publishing the information, and allowing the administrator user to upload the data (Figure 1).
B. Diagrams and Models
UML diagrams (use cases and sequence) were used to represent user requests and the interaction of system components. The information storage structure was designed using the PostgreSQL database management system to allow user management, data hosting and massive uploads of information on the platform product of the collection of specimens, taking as reference the structure of the Darwin Core standard used by the SiB Colombia and the Humboldt Institute.
C. Platform Modules
The four modules proposed for the BiolCol platform were built: user access control, user administration, information registration and consultation module. The platform includes images of the specimens collected in different places in the Department of Santander.
The information registration functionality for the BiolCol technological platform was developed through massive data uploads, allowing the download of a template in Excel format for data registration and later uploading the file to the system database.
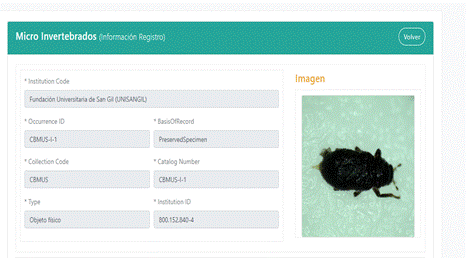
Once the data is loaded, the list of macroinvertebrates in the form can be seen, and by clicking on the icon next to the specimen code, more details about it are displayed, as can be seen in the Figure 2.
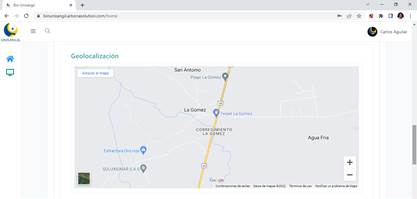
The platform allows visualizing the place where the specimen was collected, as can be seen in the Figure 3.
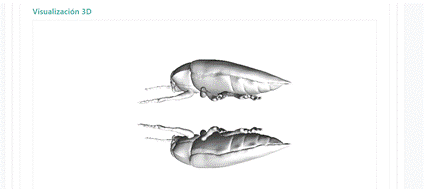
The system has the option to view the macroinvertebrates in 3D for some of the collected specimens. The different models and images are saved in a folder on the server and related to the database. Each model is related to its corresponding specimen and labels. An example of this display option can be seen in the Figure 4. The platform allows rotating the image 360 degrees to see the organism in more detail.
The fourth module of the BiolCol platform allows queries and generates labels to print and identify the organisms in the laboratory's physical collection. Researchers can search for codes associated with organisms and generate a PDF file with QR codes for printing. The QR code allows to consult the information registered on the platform from any device and send it to print. Figure 5 shows an example of the file generated to print and mark the bottles stored together with the organisms in a laboratory of the Institution.
IV. DISCUSSION
The BiolCol platform was created to give visibility to the information of the UNISANGIL CBMUS biological collection and to host information from other biological collections that use the Darwin Core standard for specimen information management. The platform allows the upload of data through an Excel template, detailed visualization of the information, 3D visualization of some specimens and generation of labels to identify the organisms in the physical collection.
Biological Collection is a bioinformatics tool that allows the registration, storage and visualization of information from the specimens of the CBMUS-UNISANGIL biological collection. The platform is accessible through the web. The registration is intuitive, friendly, agile and graphic. Users have different roles in the system and can easily access the application.
The definition of user stories was used to manage the requirements of the BiolCol application, which allowed detailed visualization of the behavior of the final product before the users. The client provided feedback during the testing phase, which led to the final approval of the software development and the successful fulfillment of the project's expectations.
The new web application allowed the digitization of specimens collected in the field more quickly, which reduced the time spent on systematizing data from the biological collection that researchers frequently update.
On many occasions, the information is not used due to the lack of technological tools and knowledge for data management. For this reason, automation and the correct use of data are necessary to generate knowledge.