Introduction
Ready to eat (RTE) meat products are popular convenience foods. In recent years, the popularity of them have increased as they are a quick, easy and accessible option for consumers 1. They are retailed chilled with a shelf life depending on their formulation, and handling. During handling, which includes slicing and packing, these products might be contaminated with microorganisms from the supermarket environment (e.g. slicer, counter, refrigerator, staff, etc)1. In addition, if the cold supply chain is abused and the products are not handled as stated by the regulations, they might be rapidly spoiled. Lactic acid bacteria (LAB) are widespread in nature and consistently exist on mucous membranes and body. Hence, they are part of the initial flora on raw meats after slaughter and in sliced cooked meats they are considered contaminants. LAB may inhibit the growth of pathogenic microorganisms as a result of the production of organic acids and other antagonist substances (2; however, organic acids might also accelerate the decomposition of meat products (3. It has been reported that LAB cause acidification, increase viscosity, excessive drip and discoloration (greening) in meat products (4 -7). Kreyenschmidt et al. (2010) reported that the shelf life of meat products ends when LAB reach 107 CFU/g, as the product is no longer sensorially acceptable because of the existance of signs of spoilage such as off-odours and viscosity 6. Mexican legislation for meat products does not required analysis of LAB; although they are of great importance since they cause economic losses during shelf life. In Mexico, most RTE meat products are sliced at supermarkets, this practice may increases the degree of contamination since one slicer is used for different products. For these reasons the aims of this study were to quantify and presumptive identify LAB strains in sliced ham as an indicator of its shelf life.
Materials and methods
Ham samples
Six samples of 250 g commercial cooked ham sliced at the retail point and elaborate without starter cultures were obtained on six consecutive Mondays at the same time from five different supermarkets of the same retail chain in the City of Chihuahua, Mexico. The study was conducted from April to December 2012. Microbiological analyses were done within the first 24h, all samples were kept under refrigeration until analyses.
Microbiological analyses
Ten grams of cooked ham were taken aseptically, placed in a stomacher bag (Lab-blender type 80 Modelo: BA 6020, USA) with ninety ml of buffered peptone water (BD Difco(, Sparks, MO, USA) for 1 minute. After decimal dilutions, samples were surface-plated (100 (l) onto appropriate media. Lactobacillus were plated on de Man Rogosa, Sharpe agar (MRS; Oxoid(, Basingstoke, Hampshire, England), mesophilic and thermophilic Streptococcus on M17 agar (Oxoid(, Basingstoke, Hampshire, England) containing 1% lactose (Sigma Aldrich, Basingstoke, Hampshire, England), Lactococcus on LM17 (BD(, Sparks Glencoe, MD, USA) containing 100 (g per liter of cycloheximide (Fluka Chemica(, Milan, Italy) and Enterococci on Kanamicin Aesculin Azide agar containing kanamycin (KAA; Oxoid, Basingstoke, Hampshire, England). Incubation was at 30(C for Lactobacillus and mesofilic Streptococcus, at 25ºC for Lactococcus and 37ºC for thermophilic Streptococcus and Enterococcus, all under anaerobic conditions (Gas-Pack System(, BBL) for 48-72h.
In order to multiply the cultures, twenty-one colonies were randomly picked and inoculated in MRS broth at 30°C for presumptive Lactococcus, Lactobacillus and Leuconostoc, and in Brain Heart Infusion broth (BHI, Lab M(, Bury, Lancashire, UK) for presumptive Enterococcus and Streptococcus at 37°C. Then colonies were transferred to nutrient agar (Oxoid(, Basingstoke, Hampshire, England) to assure purity. All strains were Gram stained and tested by the KOH method, catalase formation directly onto each plate, and morphology by phase contrast microscopy. The strains were cryopreserved in vials (Nalgene®) with liquid medium (60% glycerol (Fermont(, Monterrey, Nuevo León, México) and 40% of the strain in a liquid medium) at -20°C until they were used for characterization.
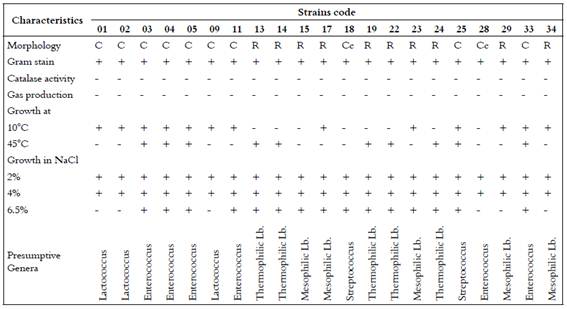
Working cultures were grown in MRS and M17 broth media for rods and cocci respectively. Overnight Gram-positive and catalase-negative strains were further tested for their ability to produce CO2 from glucose in MRS broth containing Durham tubes, growth after incubation at 10°C for 7 days, at 45°C for 2 days, and in the presence of 20, 40 and 65 g NaCl/L after incubation at 30°C for 4 days. Based on these results, all strains were presumptively classified to genus level (8), as shown in Table 1.
Table 1 Morphological and physiological criteria used in presumptive identification of the isolates in this study
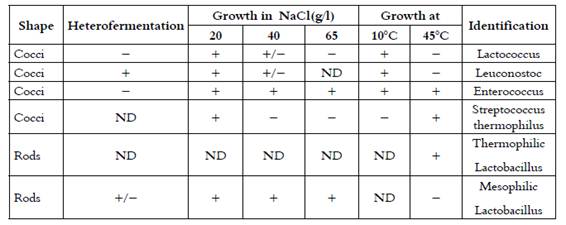
+, Positive; (, negative; +/(, positive or negative; ND, not determined.
The fermentation of carbohydrates was determined on a standard liquid media using phenol red (BD, Sparks Glencoe, MD, USA) as pH indicator. The carbon sources were Arabinose, Cellobiose, Fructose, Galactose, Glicerol, Lactose, Maltose, Mannose, Mannitol, Melibiose, Melezitose, Raffinose, Rhamnose, Ribose, Sorbitol, Sucrose, Trehalose, Xylose added to the medium to a final concentration of 1% (w/v), and Salicin, added to a final concentration of 0.5% (w/v), (all from Sigma Aldrich, Basingstoke, Hampshire, England). Aliquots of 250 µL of each media were added to a microplate (Costar, Cole-Parmer, USA) and inoculated. Incubation was microaerophyically at 35°C for 24 h. Microaerophilic conditions were generated by sealing the microplate with parafilm. The carbohydrate fermentation was considered positive when the color of the media changed to yellow. The strains were presumptively classified to genus and species level by matching results with Bergey's Manual (8.
Statistical Analysis
Bacterial counts were transformed from CFU/g to log10 CFU/g. Data were analyzed by ANOVA with significant differences (p< 0.05). Subsequently the data were subjected to analysis of variance by fitting a model that included the fixed effects of supermarket, species and their interaction using PROC GLM( (SAS, 2002). The comparison of means was performed by the Tukey-Kramer test (SAS). The isolation and identification of LAB species was reported in the number and percent of isolations and the species identified.
Results
Table 2 shows LAB counts of sliced cooked ham analyzed in this study. Differences (p<0.05) between microbial genuses were found. Enterococci showed the lowest count (2.34 ± 0.05 log10 CFU/g) and Lactobacillus the highest (5.98 ± 0.04 log10 CFU/g). No difference (p>0.05) was observed in genus between supermarkets.
Table 2 Numbers of lactic acid bacteria from cooked ham sliced at retail point estimated in different microbial media
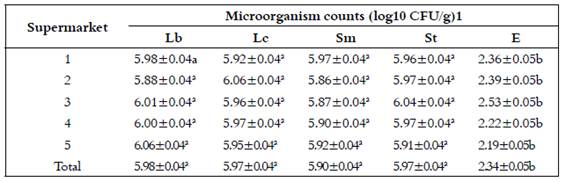
ab Means with different superscripts in the same row differ (P<0.05). Lb=Lactobacillus, Lc=Lactococcus, Sm=Streptococcus mesophiles, St= Streptococcus thermophiles and E= Enterococcus. CFU/g = colony forming units per gram. 1 Lactococcus plated onto LM17 + 100 (g/lt cycloheximide (25ºC); Streptococcus onto M17 + 1% lactose (30 and 37, for mesophiles and thermophiles, respectively), Enterococcus onto KAA agar (37ºC), all incubated under anaerobic conditions
Table 3 shows the results of the 21 isolates that were randomly picked and presumptively identified as: thermophilic Lactobacillus (23.8%), mesophilic Lactobacillus (23.8%), Enterococcus (28.6%), Lactococcus (14.3%), and Streptococcus (9.5%). Of these, only six could be characterized to species level; one was L.lactis subsp. lactis (01) and five were L. amylolyticus (isolates numbers 13, 14, 19, 22 and 24).
Discussion
Lactobacillus had the highest counts (5.98 ± 0.04 log10 CFU/g) followed by Lactococcus and Streptococcus thermophiles (5.97±0.04 log10 CFU/g), Streptococcus mesophiles (5.90±0.04 log10 CFU/g) and Enterococcus (2.34±0.05 log10 CFU/g). Meanwhile, L.lactis subsp. lactis (1 isolate) and L. amylolyticus (5 isolate) were identified. These suggest that the products have consistent quality, in terms of BAL counts, regardless of the supermarket where they were purchased. Although different supermarkets were sampled, these were from the same retail chain, so it can be assume that the cleaning and disinfection protocols are the same in all sampled supermarkets. As reported by Kreyenschmidt et al. the shelf life of RTE meat products ends when LAB counts reached 107 CFU/g (8 log10 CFU/g) 6. The results found in this study were similar to those found by Pérez-Rodríguez et al. who reported LAB counts of 5.92 ± 1.06 log10 CFU/g in cooked ham (1). Moreover, Samelis et al., at the day of purchase, reported LAB counts of 3.48 log10 CFU/g; then, at day 15, counts exceeded 8 log10 UFC/g 9. This suggests that the samples in this study might have a shelf life shorter than 15 days since at the day of purchase counts were higher than 5 log10 for all microbial genuses but Enterococci. Borch et al. found that during the aerobic storage of cooked and sliced meat products the dominant LAB were Bacillus, Lactobacillus and Micrococcus4. In cooked sausages it was reported the presence of Pediococus, Lactobacillus, Enterococcus and Aerococcus (3. Gibson et al. studied cross contamination using a melamine copolymer resin (fluorescent compound) as a contaminant, demonstrating how easily the resin was distributed. It was identified that the most likely areas of contamination were; gloves, the slicing machine, the collection tray and the blade 10. Slicing is of vital importance, as the higher the contamination of cooked meats at this stage, the shorter the shelf life, independent of the storage conditions (7,11,12.
Respect to the strains presumptive identified L.lactis subsp. lactis and L. amylolyticus, Rodríguez et al. analyzed Spanish fermented sausages samples reporting the presence of Lactococcus and isolated two strains of Lc. Lactis13. While Hamasaki et al. identified strains of Lc. lactis subsp lactis in cooked sliced ham 14. The lack of reports on the presence of Lc. lactis in meat products may be attributed to the research being addressed to dairy products given the supposition that they are found only in them, so isolates are classified as “atypical” strains. Meanwhile, L. amyloliticus have been also isolated from fresh meats (15, so that its presence in cooked ham may be due to contamination. Although phenotypic characterization, based on sugar fermentation pattern and conventional phenotypic properties, might not always provide sufficient information for the reliable identification of LAB, this is a useful tool for presumptive characterization 16. The findings in this study may reflect the reality in Mexico, since all supermarkets are handled similarly in the deli area. However, these should be viewed with caution, since the samples were taken from one supermarket chain. Therefore, it is recommended that in future works, samples of sliced cooked ham are obtained from several supermarket chains. Likewise, Mexican legislation for cooked meat products does not require analysis of LAB, just some pathogens such as Escherichia coli, Staphylococcus, and Salmonella. The results of this study show the importance of LAB in cooked ham quality and its shelf life, suggesting that it is important to regulate their content and presence, to improve food quality by regulating the handling and storage.
Conclusions
The LAB initially present in the samples showed counts between 5.98±0.04 and 2.34±0.05 at the day of purchase, being Lactobacillus the most numerous group. L.lactis subsp. lactis and L. amylolyticus were identified as a spoilage bacteria of sliced cooked ham. Meat products are popular convenience foods that are highly perishable and rapidly spoiled due to defective handling. High LAB counts are a common cause of spoilage in meat products since these are commonly found in meat environments. The quantification and phenotyphic methodologies based on sugar fermentation are helpful to better understand the growth and presumptive identification of LAB as a spoilage microorganism of sliced cooked ham. The study of the evolution of the LAB microbiota in sliced cooked ham may be important for selecting the main deteriorating LAB aiming to decrease their presence in this food product. Therefore a study on the processing, distribution and in-site handling of RTE meat products may be necessary in order to increase their shelf life. Also, the evaluation might include enumeration and growth rate of LAB and the presence and quantification of organic acids and other chemical compounds.
Acknowledgements
This research was supported by the Professional Development Program for Teachers (Programa para el Desarrollo Profesional Docente, PRODEP). The authors declare no conflict of interest