INTRODUCTION
Staphylococci are present as normal flora on the skin and anterior nares of humans, and up to two-thirds of the population are estimated to be colonized by it. They are also responsible for causing skin and soft tissue infection to fatal endovascular infections and sepsis. In recent years, the importance of staphylococci has increased because of their causative role in nosocomial infections in both developed and developing countries (1-3). The widespread use of immunosuppressive drugs, indwelling intravascular catheters, artificial tools, and health care settings have been reported to be associated with the spread of these infections. The occurrence of bacterial infection had decreased with the discovery of penicillin in 1940 until S. aureus began producing β lactamase. The resistance to penicillin drove the development of methicillin drugs in 1960. But the first strain of methicillin-resistant S. aureus (MRSA) was reported in 1961, and since then, the infection caused by MRSA has been a major public health concern (4,5). The dissemination of MRSA has developed in parallel with increasing the prevalence of multi drug resistant strains (MDR).
One major virulence property of staphylococci is its ability to form biofilm. Biofilms are communities of microorganisms embedded in the extracellular polymeric substances (EPS) matrix. Bacterial biofilms provide considerable advantages like protection against antimicrobial agents, acquisition of new genetic traits, and resistance against phagocytosis and other components of the body defense system (6). Although vancomycin has been the most reliable therapeutic agent against infections caused by MRSA, an increasing number of vancomycin intermediates sensitive to S. aureus (VISA) are being reported. However, the vancomycin-resistant S. aureus (VRSA) is still limited worldwide (5,7). In addition, emerging data suggest that vancomycin may be less effective against severe MRSA infection with minimum inhibitory concentration (MIC) values at the higher end of the susceptibility range designated by Clinical and Laboratory Standards Institute (CLSI), resulting in treatment failure and increased mortality. Many researchers have reported that the higher MIC of vancomycin to MRSA, even in the susceptible range, correlates with the drug- resistant to different classes of antibiotics (7-9). Only limited data are available on the susceptibility of MRSA to vancomycin in Nepal. Although VRSA has not been reported from Nepal, increasing trends of decreased susceptibility of MRSA to vancomycin is reported indicating the need for more research in this field (6,8,10). With the increasing occurrence of MRSA having decreased susceptibility to vancomycin and biofilm formation, the choice of medication remains one of the most challenging concerns in the management of infection caused by such isolates. Thus, this study was conducted to determine the changing susceptibility pattern of methicillin-resistant staphylococci isolates to vancomycin and biofilm producing capacity of S. aureus and coagulase negative staphylococci (CNS) isolated from different clinical specimens.
MATERIALS AND METHODS
Isolation and identification of staphylococci
The hospital-based cross-sectional descriptive study was conducted at the Department of Microbiology, B & B hospital, Gwarko, and KIST Medical College and Teaching Hospital, Imadol, Lalitpur, Nepal, from January 2018 to December 2018. 375 staphylococci were isolated from different clinical specimens processed during the study period. Specimens processed were wound/pus (w/s), blood, urine, body fluids, various tips (central venous catheter, catheter, endotracheal, transtracheal), etc. All clinical samples were collected and processed following standard microbiological procedures. The isolated colonies were identified by inoculating in Mannitol salt agar media, Blood agar media and performing Gram staining and specific biochemical tests. The slide and tube coagulase tests were performed to differentiate S. aureus and CNS (11). The species of CNS were identified based on a simplified scheme proposed by Cunha et al. using several biochemical tests (12). The biochemical tests performed were clumping factor, hemolysis, pyrrolidonyl arylamidase, urease, alkaline phosphatase, nitrate reduction, acetoin production, acid production from sucrose, mannose, trehalose, xylose, and susceptibility to novobiocin and desferrioxamine.
Antimicrobial susceptibility test
The modified Kirby-Bauer disk diffusion method was carried out to determine in vitro antibiotic susceptibility to nine antimicrobial agents selected based on different modes of action. Antibiotic discs (HiMedia, India) used was ciprofloxacin (5µg), clindamycin (2µg), erythromycin (15µg), gentamicin (10µg), penicillin (10 units), cotrimoxazole (1. 25 / 23 / 75 µg), tet r ac ycline ( 3 0 µg) and chloramphenicol (30µg). The test used a bacterial suspension with turbidity adjusted equivalent to a 0.5 McFarland standard, and performed on Mueller Hinton agar plates (HiMedia, India) based on the Clinical & Laboratory Standards Institute (CLSI) guidelines (13). S. aureus ATCC 25923 strain was used as a positive control. Screening for Methicillin resistant staphylococci was carried out by cefoxitin disc diffusion method and interpreted according to CLSI guidelines. The growth of staphylococci with a zone of inhibition around cefoxitin disc (ZOI) ≥ 22mm was identified as methicillin sensitive S. aureus (MSSA), and that of ZOI ≤ 21 was identified as methicillin resistant S. aureus (MRSA). Similarly, for coagulase-negative staphylococci, ZOI ≥ 25 were identified as methicillin sensitive coagulase negative staphylococci (MSCNS) and ZOI ≤ 24 as methicillin resistant coagulase negative staphylococci (MRCNS), respectively.
Determination of minimum in hibitory concentration
Minimum inhibitory concentration (MIC) technique by agar dilution method was performed to determine the vancomycin-intermediate and resistant and oxacillin resistant strains of staphylococci isolate. MIC to vancomycin and oxacillin of all isolates was done using the agar dilution method following CLSI guidelines (14). Different concentrations of 0.125 to 32μg/ml of vancomycin and 0.125μg/ml to 128μg/ml of oxacillin incorporated plates were prepared. Positive growth controls were kept for each isolate, and S. aureus (ATCC 29213) of known MIC was included in each test to control antibiotic potency. All the experiments were performed in triplicate.
Detection of biofilm formation
The Tissue Culture Plate (TCP) method was used to determine the biofilm production of Staphylococcal isolates as described by Christensen et al. with some modifications (15,16). In this assay, a loopful of organisms was inoculated in 5mL Tryptone Soybroth (TSB) (HiMedia Laboratories Private Limited, India) supplemented with glucose and incubated at 37ºC for 24 hours. The overnight culture was diluted 1:100 with fresh media, and 0.2mL of this diluted culture was then inoculated into individual wells of sterile polystyrene 96 well flat bottom tissue culture plates and incubated at 37ºC for 24 hours. After incubation, the content of the tissue culture plate was removed by gentle tapping and washed with PBS (pH 7.2) 4 times to remove free-flowing planktonic bacteria. Biofilms formed by adherent sessile bacteria in the plate were fixed with sodium acetate (2%). It was then stained with crystal violet (0.1%) for 15 minutes at room temperature. Excess stain was rinsed off by washing with deionized water 4 times, and plates were dried. The optical density (OD) of stained adherent bacteria was measured with a micro ELISA auto reader at 630nm (OD 630nm). OD values from sterile medium, fixative, and dye were averaged and subtracted from all test values. Experiments were performed in triplicates and repeated three times. Bacterial adherence was classified based on OD values of the individual isolates: < 0.12, 0.12 - 0.24, and > 0.24 as weak, moderate, and strong biofilm formation, respectively.
Detection of ica gene by PCR
The biofilm of staphylococci is composed of the layer of extracellular polymeric substance called polysaccharide intercellular adhesion (PIA) encoded by ica operon (icaADBC genes). The genomic DNA was extracted from all 375 staphylococcal isolates as previously described (16) using the DNA extraction kit, following the manufacturer’s instructions (Thermo Fisher Scientific Inc.). The sequences of icaA and icaD (accession number U43366) were taken from the GenBank sequence of the National Center for Biotechnology Information (NCBI) database. Primers (Solis Biodyne, Denmark) specific for icaA (forward 5’-TCTCTTGCAGGAGCAATCAA and reverse 5’-TCAGGCACTAACATCCAGCA) generating a product size of 188bp and icaD (forward 5’-ATGGTCAAGCCCAGACAGAG and reverse 5’-CGTGTTTTCAACATTTAATGCAA) with the product size of 198bp were designed by the Primer3 program. The PCR product was analyzed in 2% agarose gel stained with SYBR safe dye (Invitrogen).
RESULTS
375 staphylococci were isolated from different clinical specimens collected at two tertiary care hospitals in Nepal, comprising 42.9 % S. aureus and 57.1 % coagulase-negative staphylococci (CNS). S. epidermidis (57.5 %) was the most commonly isolated CNS, followed by S. saprophyticus (18.7 %), S. haemolyticus (11.2 %), S. hominis (7.0 %), and S. capitis (5.6 %).
Distribution of methicillin resistant staphylococci
Out of 161 S. aureus, 81.4 % isolates were MRSA and 18.6% MSSA as detected by disc diffusion assay. Similarly, 66.8% represented MRCNS of 214 CNS isolates. The highest rate of resistance of MRSA isolates was against penicillin (96.2 %), followed by erythromycin (84 %). A similar resistant pattern was observed in MRCNS, with 96.5 % and 84.6 % isolates resistant to penicillin and erythromycin, respectively. All MSSA isolates were sensitive to clindamycin and chloramphenicol. Similarly, the highest resistance was seen towards penicillin for both MSSA (90 %) and MSCNS (81.7 %) (Table1).
Table 1 Antibiotic susceptibility pattern of methicillin resistant and methicillin sensitive staphylococci
| Antibiotics | Potency (µg/disc) | MRSA (n=131) | MSSA (n=30) | p value | MRCNS (n=143) | MSCNS (n=71) | p value |
|---|---|---|---|---|---|---|---|
| Penicillin | 10 units | 126 (96.2%) | 27 (90.0%) | 0.160 | 138 (96.5%) | 58 (81.7%) | 0.000 |
| Ciprofloxacin | 5 | 102 (77.9%) | 17 (56.7%) | 0.017 | 69 (48.3%) | 7 (9.9%) | 0.000 |
| Tetracycline | 30 | 8 (6.1%) | 1 (3.3%) | 0.551 | 22 (15.4%) | 5 (7.0%) | 0.084 |
| Clindamycin | 2 | 16 (12.2%) | - | 0.044 | 53 (37.1%) | 10 (14.1%) | 0.001 |
| Chloramphenicol | 30 | 4 (3.1%) | - | 0.332 | 13 (9.1%) | 6 (8.5%) | 0.877 |
| Erythromycin | 15 | 110 (84.0%) | 17 (56.7%) | 0.001 | 121 (84.6%) | 34 (47.9%) | 0.000 |
| Cotrimoxazole | 1.25/23.75 | 72 (55.0%) | 18 (60.0%) | 0.616 | 65 (45.5%) | 15 (21.1%) | 0.001 |
| Gentamicin | 10 | 58 (44.3%) | 9 (30.0%) | 0.152 | 42 (29.4%) | 3 (4.2%) | 0.000 |
MRSA= Methicillin resistant Staphylococcus aureus, MSSA= Methicillin sensitive Staphylococcus aureus, MRCNS= Methicillin resistant coagulase negative staphylo- cocci, MSCNS= Methicillin sensitive coagulase negative staphylococci
Distribution of oxacillin and vancomycin MICs in Staphylococcal isolates
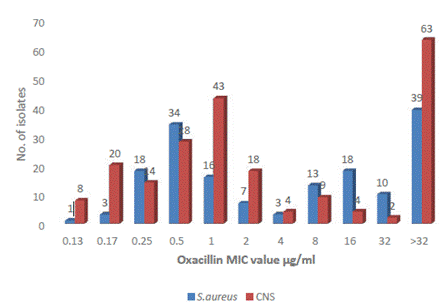
The staphylococcal isolates had MIC values for oxacillin from 0.125 to >32 µg/ml. According to the observed MIC values, there were 83 MRSA (MIC ≥4µg/ml) isolates and 171 MRCNS (MIC ≥0.5 µg/ml) isolates as per CLSI. S. aureus ATCC 29213 recorded MIC of 0.5 µg/ml used as positive control strain. Sixty-three CNS isolates and 39 S. aureus had MIC >32 µg/ml (Figure 1).
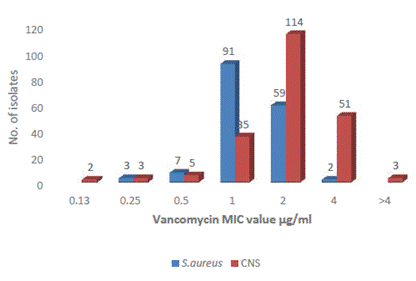
Most S. aureus strains were sensitive to vancomycin and the MIC value recorded was 0.125 µg/ml to ≤2 μg/ml, where 59 isolates had MIC value at a clinical breakpoint of 2 µg/ml. The two S. aureus isolates had a MIC value of 4 μg/ml; however, these are not resistant as per CLSI. These two isolates with intermediate susceptibility to vancomycin were obtained from central venous catheter (CVC) and catheter samples of inpatients. Similarly, among CNS also, all isolates were sensitive with a MIC of ≤4 μg/ml, whereas 51 CNS had MIC at a clinical breakpoint of 4 µg/ml. Three isolates had MIC values >4 μg/ml representing intermediate susceptibility to vancomycin. These 3 isolates were methicillin-resistant and had been isolated from blood, CVC, and wound/pus (w/p) (Figure 2). S. aureus ATCC 29213 recorded a MIC of 1 μg/ml used as control strain.
Distribution of oxacillin MIC in methicillin resistant isolates
Fifty-five isolates detected by cefoxitin disc diffusion assay as MRSA were susceptible to oxacillin by agar dilution method, whereas 7 MSSA isolates were resistant to oxacillin. Similarly, only 20 CNS isolates were detected as methicillin-resistant by disc diffusion assay and were susceptible to oxacillin, but 49 CNS isolates sensitive to methicillin were resistant to oxacillin (Table 2).
Table 2 MIC of oxacillin in methicillin resistant and methicillin sensitive staphylococci
| MIC value of oxacillin (µg/ml) | MRSA (n,%) | MSSA (n,%) | MRCNS (n,%) | MSCNS (n,%) |
|---|---|---|---|---|
| 0.125 | 3 (2.3%) | - | 14 (9.8%) | 15 (21.1%) |
| 0.25 | 12 (9.2%) | 7 (23.3%) | 6 (4.2%) | 7 (9.9%) |
| 0.5 | 24 (18.3%) | 9 (30.0%) | 22 (15.4%) | 6 (8.5%) |
| 1 | 11 (8.4%) | 4 (13.3%) | 32 (22.4%) | 15 (21.1%) |
| 2 | 5 (3.8%) | 3 (10.0%) | 11 (7.7%) | 7 (9.9%) |
| 4 | 3 (2.3%) | - | 4 (2.8%) | 1 (1.4%) |
| 8 | - | 1 (3.3%) | 6 (4.2%) | 2 (2.8%) |
| 16 | 10 (7.6%) | 2 (6.7%) | 4 (2.8%) | - |
| 32 | 17 (13.0%) | 1 (3.3%) | 2 (1.4%) | - |
| ˃32 | 46 (35.1%) | 3 (10.0%) | 42 (29.4%) | (25.4%) |
Distribution of vancomycin MIC in methicillin resistant isolates
All staphylococcal isolates were sensitive to vancomycin irrespective of methicillin resistance. The MRSA had vancomycin MIC values from 0.25 to 4 µg/ml. Forty-nine MRSA isolates and 8 MSSA isolates had MIC values at a clinical breakpoint of 2 µg/ml. Three MRSA were detected with intermediate susceptibility to vancomycin. All CNS were also sensitive to vancomycin, but 34 MRCNS and 20 MSCNS had a clinical breakpoint of 4 µg/ml (Table 3).
Table 3 MIC of vancomycin in methicillin resistant and methicillin sensitive staphylococci
| MIC value of vancomycin (μg/ml) | MRSA (n,%) | MSSA (n,%) | MRCNS (n,%) | MSCNS (n,%) |
| 0.125 | - | - | 1 (0.7%) | 1 (1.4%) |
| 0.25 | 2 (1.5%) | 2 (6.7%) | - | 2 (2.8%) |
| 0.5 | 4 (3.1%) | 3 (10.0%) | 5 (3.5%) | - |
| 1 | 73 (55.7%) | 17 (56.7%) | 27 (18.9%) | 8 (11.3%) |
| 2 | 49 (37.4%) | 8 (26.7%) | 76 (53.1%) | 40 (56.3%) |
| 4 | 3 (2.3%) | - | 34 (23.8%) | 20 (28.2%) |
Biofilm formation in staphylococci isolates
Biofilm formation detected by the TCP method showed that 22.1 % of staphylococci isolates were strong biofilm producers with 24.1 % MRSA and 9.3 % MRCNS. Similarly, the ica genes were detected among 22.9 % of staphylococcal isolates, where 18 % and 13.1 % represented MRSA and MRCNS, respectively (Table 4).
Table 4 Detection of biofilm formation by different methods among methicillin resistant and methicillin sensitive staphylococci
| Method | Biofilm formation | MRSA | MSSA | P value | MRCNS | MSCNS | P value | Total |
|---|---|---|---|---|---|---|---|---|
| TCP method | Strong | 39 (24.2%) | 9 (5.6%) | 0.681 | 20 (9.3%) | 15 (7%) | 0.412 | 83 (22.1%) |
| Moderate | 31 (19.3%) | 5 (3.1%) | 38 (17.8%) | 17 (7.9%) | 91 (24.3%) | |||
| Weak/Non | 61 (37.9%) | 16 (9.9%) | 85 (39.7%) | 39 (18.2%) | 201 (53.6%) | |||
| ica gene | Presence | 29 (18%) | 16 (9.9%) | 0.001 | 28 (13.1%) | 13 (6.1%) | 0.824 | 86 (22.9%) |
| Absence | 102 (63.4%) | 14 (8.7%) | 115 (53.7%) | 58 (27.1%) | 289 (77.1%) |
MIC of oxacillin in biofilm producing staphylococci
Out of 174 staphylococci biofilm producers detected by the TCP method, 48.3 % were S. aureus and 51.7 % CNS. Among them, 53.6 % isolates were MRSA (4 µg/ml), and 84.4 % were MRCNS (0.5 µg/ml). The ica genes were detected in 86 isolates with, 52.3 % and 47.7 % isolates being S. aureus and 41 CNS, respectively. 44.4 % and 73.2 % of isolates were detected as MRSA and MRCNS by the agar dilution method (Table 5).
Among all staphylococci positive for biofilm production by the TCP method, only one was intermediate susceptible to vancomycin. This isolate was S. aureus obtained from CVP, which was also resistant to oxacillin. The other two isolates that were intermediately susceptible to vancomycin were non- biofilm producers and were isolated from catheters and w/p. The ica genes were absent in these three isolates (Table 5).
Table 5 MIC of oxacillin and vancomycin in biofilm producing staphylococci
| Aspect | No of biofilm producer S. aureus isolates determined by | No of biofilm producer CNS isolates determined by | ||
| TCP method | Ica genes | TCP method | Ica genes | |
| Oxacillin MIC (μg/ml) | ||||
| 0.125 | 1 | 1 | 8 | 6 |
| 0.25 | 13 | 9 | 6 | 5 |
| 0.5 | 15 | 7 | 14 | 6 |
| 1 | 6 | 3 | 16 | 5 |
| 2 | 4 | 5 | 14 | 4 |
| 4 | 3 | 2 | 3 | 3 |
| 8 | 6 | 5 | 3 | 1 |
| 16 | 10 | 6 | 2 | - |
| 32 | 4 | 5 | 1 | 2 |
| 64 | 22 | 2 | 23 | 9 |
| Vancomyci n MIC (µg/ml) | ||||
| 0.125 | - | - | - | 1 |
| 0.25 | 2 | 1 | 1 | 1 |
| 0.5 | 2 | 3 | 3 | 1 |
| 1 | 47 | 26 | 19 | 5 |
| 2 | 32 | 15 | 48 | 24 |
| 4 | 1 | - | 19 | 9 |
Distribution of MIC of oxacillin and vancomycin in different species of CNS
The agar dilution method detected 41.6 % S. epidermidis, 16.4 % S. saprophyticus, 11.2 % S. haemolyticus, 4.7 % S. capitis, and 6.5 % S. hominis resistant to methicillin.
All CNS species were susceptible to vancomycin. The MIC ranged from 0.125 µg/ml to 4 µg/ml for S. epidermidis and S. saprophytics, while for S. haemolyticus, the range was 0.5 µg/ml to 4 µg/ml, and for S. capitis and S. hominis, the MIC range was 1 to 4 µg/ml (Table 6).
Table 6 MIC of oxacillin and vancomycin in different species of CNS
| Species of CNS | MIC of oxacillin (µg/ml) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ˃32 | Total | |
| S. epidermidis | 22 | 12 | 17 | 28 | 9 | 1 | 5 | 1 | - | 28 | 123 |
| S. saprophyticus | 4 | 1 | 7 | 6 | 3 | 1 | 1 | 2 | - | 15 | 40 |
| S. haemolyticus | - | - | 2 | 10 | 2 | - | 1 | 1 | 1 | 7 | 24 |
| S. capitis | 2 | - | 1 | 1 | 1 | 1 | 1 | - | - | 5 | 12 |
| S. hominis | 1 | - | 1 | 2 | 3 | 2 | - | - | 1 | 5 | 15 |
| MIC of vancomycin | |||||||||||
| S. epidermidis | 1 | 2 | 3 | 20 | 67 | 30 | - | - | - | - | 123 |
| S. saprophyticus | 1 | - | 1 | 6 | 21 | 11 | - | - | - | - | 40 |
| S. haemolyticus | - | - | 1 | 5 | 11 | 7 | - | - | - | - | 24 |
| S. capitis | - | - | - | 2 | 8 | 2 | - | - | - | - | 12 |
| S. hominis | - | - | - | 2 | 9 | 4 | - | - | - | - | 15 |
DISCUSSION
In recent years, staphylococcal isolates have become one of the most dangerous pathogens due to their increased resistance to various commonly used antibiotics and biofilm formation. Studies showed that MRSA and MRCNS are the causative agent of hospital-acquired infection and incipient community pathogens in many geographical regions (6,8).
In our study, 81.4 % and 66.8 % of isolates were MRSA and MRCNS by cefoxitin disc diffusion assay, higher than in other studies from Nepal, where MRSA representation ranged from 19 % to 45.9 % (17-22). Higher isolation rates reported in our study can be attributed to several factors. Some of these include indiscriminate use of antibiotics, population and area studied, lack of awareness, and failure to observe simple yet effective infection control precautions like strict patient isolation and frequent hand washing by health care personnel (23).
Most MRSA and MRCNS isolates were sensitive to tetracycline and chloramphenicol, indicating exposure to newer drugs and susceptibility to older drugs. Most of these isolates were observed to be resistant to penicillin and erythromycin. This discrepancy suggests a change in antibiotic susceptibility pattern with a difference in study settings, geographical location, and population studied. This resistance pattern was in concordance with other studies (23). The notoriety of staphylococci in developing resistance to therapeutic agents has been known since the advent of penicillin resistance, particularly in response to selective antibiotic pressure.
The MIC value of S. aureus for oxacillin showed that 55 MRSA determined by the cefoxitin disc diffusion method were, in reality, susceptible to oxacillin. The MIC of MSSA isolates agreed with the methicillin status except for 7 MSSA isolates with MICs ≥ 4 µg/ml. For CNS, 20 methicillin-resistant isolates determined by the cefoxitin disc diffusion method were susceptible to oxacillin, but 49 methicillin- sensitive CNS were characterized as oxacillin resistant. Such elevated MICs are obtained due to mutation in one of their Penicillin Binding Protein (PBPs) (24) or hyperproduction of staphylococcal β lactamase (25) that could explain the observed aberration. This result in phenotypes expressing oxacillin MICs between the clinical breakpoint of susceptibility and resistance that are often termed Borderline oxacillin resistant S. aureus (BORSA) that lack the mecA gene (25). Oxacillin MIC determination is limited to research purposes only. In contrast, for diagnostic purposes, the cefoxitin screening result is extrapolated for penicillinase-resistant penicillin’s susceptibility to decide if they can be prescribed to patients (13). Due to limited treatment options for infections caused by methicillin-resistant staphylococci, the treatment of such infections is often difficult leading to a prolonged hospital stay and sometimes leading to treatment failure resulting in fatal outcomes (20).
Vancomycin remains the first choice parenteral agent for both empiric and definitive therapy of methicillin-resistant staphylococcal infections. There is limited literature regarding the MIC of vancomycin for staphylococci isolated from clinical samples in Nepal. All staphylococci isolates in the present study were sensitive to vancomycin, which was in concordance with the observations in clinical practices since resistance to the last resort drug is still rare. This result is corroborated by other studies conducted on different samples (6,26-28). Until now, staphylococcal isolates resistant to vancomycin has not been reported from Nepal (28). In the present study, two S. aureus and three CNS were detected with intermediate susceptible to vancomycin. Many other authors have reported VISA and VRSA from different countries (26). This reduced susceptibility to vancomycin is attributed to exposure of the staphylococci to this antibiotic.
In our study, all CNS isolates were found sensitive to vancomycin regardless of oxacillin sensitivity, whereas three S. aureus resistant to oxacillin had intermediate sensitivity to vancomycin. Pahadi et al. (2014) have also reported four VISA isolates with MICs of vancomycin to MRSA ranging from 0.5 μg/ ml to 4 μg/ml. In a study carried out by Amatya et al. (2014), none of the MRSA isolates were reported to be VISA or VRSA but showed higher MIC (0.5 μg/ ml to 2 μg/ml) of vancomycin. The haphazard use of this antibiotic causes selective pressure resulting in the emergence of VISA/VRSA. The use of antibiotics like vancomycin has a major impact on the genetic evolution of staphylococci during the development of drug resistance (9). As vancomycin is a reserve drug, it will be challenging to treat the infections caused by VRSA, and it is emerging as a serious public health problem worldwide (6).
In the present study, reduced susceptibility to vancomycin was associated with methicillin resistance detected by phenotypic methods. Several studies indicated that vancomycin heteroresistant staphylococci were significantly related to the isolation of methicillin-resistant staphylococci among patients with bacteremia (29,30). Inadequate and inaccurate vancomycin therapy for treating infections caused by methicillin-resistant staphylococci increased the selective antibiotic pressure resulting in the selection of non-susceptible strains like VISA or heterogenous vancomycin intermediate S. aureus (hVISA) or those with elevated MICs levels. The mechanism underlying the association between methicillin resistance and reduced vancomycin susceptibility remains unclear.
In our study, the resistance of CNS to oxacillin was 80.4 % which is in accordance with other studies (30,31). Irrespective of oxacillin susceptibility, all CNS isolates in the present study were sensitive to vancomycin. It agreed with other studies conducted on different samples that demonstrated all isolates of CNS being sensitive to vancomycin (32,33). Vancomycin resistance is still infrequent in CNS. However, heterogeneous resistance was reported among CNS and was associated with vancomycin therapy failure, which may be a precursor for developing vancomycin-resistant strains (5,6). These heterogeneous resistant strains have a susceptible vancomycin MIC, but they can grow in the presence of 4 µg/mL vancomycin, which is greater than their MIC (33).
Only 83 (22.1 %) isolates were detected to be strong biofilm producers by the TCP method, and among them, 48 (57.8 %) S. aureus and 35 (42.2 %) CNS comprised biofilm producers. The biofilm producers were observed more in methicillin-resistant isolates of S. aureus and CNS. The polysaccharide layer of biofilm formation was attributed to the ica gene. The ica genes were observed among 86 (22.9 %) staphylococcal isolates distributed among 45 (28%) S. aureus and 41 (19.2 %) CNS. Most ica gene- positive isolates were also found to be methicillin- resistant. A study conducted by Mirani et al. (2013) reported that 57 % of MRSA isolates as biofilm producers. The regulation of the ica operon is complex, and the ica genes are expressed in a stressful environment, and many factors such as high osmolarity, anaerobic condition, increased temperature, and subinhibitory presence of some antibiotics contribute to its expression (34).
About 50% (45/86) of biofilm-producing S. aureus and more than half of the biofilm-producing CNS isolates (76/90) were resistant to oxacillin. Furthermore, several studies have demonstrated that methicillin susceptibility influences biofilm production and thereby the presence of ica genes (35-37). In a biofilm, the frequency of horizontal plasmid transfer is much higher than between planktonic cells. Studies on staphylococcal biofilm showed that biofilm promotes the spread of plasmid- borne antibiotic resistance genes by conjugation/ mobilization (38). This explains the acquisition of resistant genes to ica gene-positive strains (39,40).
The selective pressure generated due to the frequent use of vancomycin to treat methicillin- resistant staphylococcal infections has resulted in the emergence of resistant isolates or reduced susceptibility. In microbial communities growing in biofilm, vancomycin has a reduced effect due to the slow growth rate of bacteria and structured heterogeneity with biofilm (41). In the present study, one biofilm-producing S. aureus was detected as VISA, and 32 isolates with a clinical breakpoint of susceptibility, whereas 15 ica gene positive- isolates were detected with a clinical breakpoint of susceptibility. In the case of CNS, 19 biofilm producers and 9 ica positive isolates were found to have a clinical breakpoint of susceptibility. The emergence of many isolates with a clinical breakpoint of susceptibility to vancomycin can lead to treatment failure (39).
In our study, 89 (41.6 %) S. epidermidis were found resistant to oxacillin, followed by S. saprophyticus (35, 16.4 %). All CNS species were susceptible to vancomycin irrespective of oxacillin resistance, according to other studies (32,33,42). The study showed that although all CNS isolates were sensitive to vancomycin, isolates had a MIC of vancomycin with a clinical breakpoint of susceptibility. The presence of subpopulations with non-susceptible strains like VISA/hVISA or those with elevated MIC demonstrates the heterogeneity of CNS strains in terms of susceptibility to this antibiotic. Such resistance is endogenous (i.e., chromosomal mutation), and the mechanism is highly complex, often linking to a thickening of the bacterial cell wall, with hyperproduction of glycopeptide binding targets and altered autolysis. Additional tests are required to detect the main genes responsible for vancomycin resistance (vanA and vanB) (9). However, further investigations are required to elucidate the underlying mechanism of how these events occur in staphylococci.
Most of the oxacillin-resistant S. aureus and CNS were isolated from tips (11/18) and (45/52), respectively, indicating significant oxacillin resistance with the specimen. Staphylococci being commensal to the skin, easily gain access to the site of skin puncture and deep cuts, which cause uncomplicated infections but at times may develop into complicated infections to systemic failure. Device-related infections by staphylococci are a major problem in health care settings due to the propensity of biofilm formation by organisms that are difficult to eradicate and often lead to systemic, persistent infection and mortality (9,43,44).
The fact that the use of antimicrobial agents has a substantial impact on the genetic evolution of staphylococci during the development of drug resistance, like in the case of vancomycin, should not be ignored (44). The major drawback of this study was the lack of molecular characterization of the isolates and detection of virulent genes, which could be investigated in future research. Such a study may help physicians generate new treatment policies and develop new drugs against the resistant properties of isolates.
CONCLUSION
A very high number of methicillin-resistant isolates were observed among S. aureus and CNS. Determination of MIC showed vancomycin can still be used as the drug of choice to treat infections caused by methicillin-resistant staphylococci despite the detection of very few isolates with intermediate susceptibility. The present study demonstrated an association between the presence of the icaADBC operon, biofilm formation, and antimicrobial resistance in staphylococci isolated from various specimens. Furthermore, the detection of staphylococci subpopulations with intermediate vancomycin resistance associated with methicillin resistance highlights the role of this organism as an important multidrug-resistant microorganism and the consequent need to implement measures for the control of antibiotic use.