Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista MVZ Córdoba
Print version ISSN 0122-0268On-line version ISSN 1909-0544
Rev.MVZ Cordoba vol.15 no.3 Córdoba Sept./Dec. 2010
SHORT COMMUNICATION
Preliminary studies of the genetic structure of “Cimarron uruguayo” dog using microsatellite markers
Estudios preliminares de la estructura genética del perro cimarrón uruguayo usando microsatélites
1Universidad de la República, Facultad de Veterinaria, Área Genética. Lasplaces 1550. C.P. 11600, Montevideo, Uruguay.
2Universidad de Zaragoza, Facultad de Veterinaria, Laboratorio de Citogenética y Genética Molecular. Zaragoza, C. P. 50013, España.
*Corresponding: rgagliar@gmail.com
Recibido: Abril 12 de 2010; Aceptado: Agosto 30 de 2010.
Abstract
Objetive. To analyze the population structure, using microsatellite markers in a sample of “Cimarron Uruguayo” dogs. Materials and methods. Thirty dogs were analyzed in different areas of Uruguay with a set of nine molecular microsatellite markers using PCR. The population structure was analyzed using the free distribution software “Structure”. Results. According to our data, the preliminary results show that it is not possible to establish a subdivision among the animals in the sample. Conclusions. The study supports the hypothesis that the currently existing canines derive from a founding nucleus that took refuge in the Northeastern region of the country. The distribution of the breed among the different areas of Uruguay continues nowadays, so there is no isolation among the different groups of animals, and the exchange is constant.
Key words: Population structure; DNA markers; Cimarron Uruguayo.
Resumen
Objetivo. Analizar la estructura poblacional en una muestra de perros “Cimarrón Uruguayo” usando marcadores moleculares tipo microsatélites. Materiales y métodos. Se analizaron treinta caninos de diferentes zonas de Uruguay con un set de nueve marcadores moleculares microsatélites empleando PCR. La estructura poblacional se analizó con el software de distribución libre “Structure”. Resultados. Según nuestros datos, los resultados preliminares muestran que no es posible establecer una subdivisión entre los animales de la muestra. Conclusiones. El estudio realizado apoya la hipótesis de que los perros que existen en la actualidad derivan del núcleo fundador que se refugió en la región noreste del país. La distribución de la raza entre las distintas áreas de Uruguay continúa hoy en día, no existe aislamiento entre los diferentes grupos de animales y el intercambio es constante.
Palabras clave: Estructura poblacional, marcadores de ADN, Cimarrón Uruguayo.
Introduction
The “Cimarron Uruguayo” is the only dog breed which is originary from Uruguay. On 21 February 2006, the “Fédération Cynologique Internationale” recognizes the breed in primary form, which means that it is in a proof period of five years before its final recognition (1). This has led to an increase in the interest in the breed on the part of the breeders and the exhibitors, which is why strict controls are being made to ensure the standardization and prevent the occurrence of undesirable hereditary characteristics. At different times in history, these animals have represented a serious danger to man and livestock. Some historians, in the XVIth century, talked about their multiplication and the damage they produce, even putting a price on their heads.
In 1788, the extermination of these dogs was approved by Buenos Aires Council. During the XIXth century, the persecution of these dogs still continued. The second time when these dogs represented a problem was during the “Guerra Grande”. At this time, there was another pursuit of Cimarron dogs. However, a few individuals still remained in rural places of the provinces of Cerro Largo and Treinta y Tres (2-3). Characterization of the population structure of this dog breed is useful in a variety of contexts. Genetic ascertainment of within-species population structure has been widely applied for classifying subspecies, for defining intraspecific conservation units, for understanding events in the history of a species, for identifying ongoing speciation events and for testing hypothesis about evolutionary processes (4). “Structure” is one of the freeware used to analyze population structure. It uses multi-locus genotypic data, so it can be applied to the majority of the commonly-used genetic markers (5). Microsatellites are among the plausible genetic markers to be used for these analyses. It is assumed that there is no linkage among the chosen loci.
Having in consideration its history and the interest this breed has arisen, the objective of this investigation was to analyze the population structure, using microsatellite markers in a sample of “Cimarron Uruguayo” dogs.
Materials and methods
Animals and study area. Thirty “Cimarron Uruguayo” dogs were analyzed: 13 animals from the northeastern (NE) area of Uruguay and 17 from the southern (S) area. These areas were selected taking into account: the background of the founding nucleus in the origins of the breed (NE area: provinces of Cerro Largo and Treinta y Tres) and animal density (S area: provinces of Montevideo and Canelones).
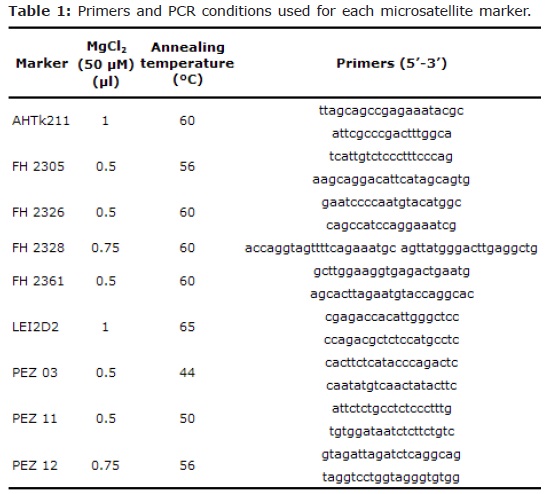
DNA isolation. DNA was isolated using the AxyPrep Multisource Genomic DNA Miniprep Kit (Axygen biosciences). Nine microsatellite markers tested in previous studies in this breed (6), included in ISAG (International Society for Animal Genetics) panel 2 were analyzed using PCR technique; each of them was amplified separately. The amplifications were performed using standard programmes (denaturation, 95°C, 4 minutes; 35 cycles of 95°C, 1 minute, 44-60°C, 30 seconds, 72°C, 45 seconds). A mix was made with the amplifications of each animal. In table 1 are shown microsatellite markers used in this work, its respective primers and PCR conditions.
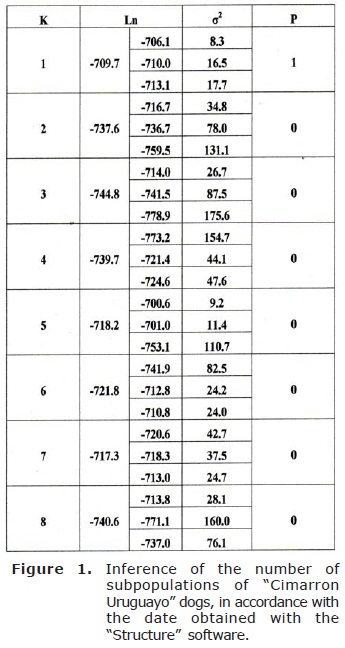
Data analysis. The results were read with the MegaBACE 1000 programme, with the permit of the University of Zaragoza (UNIZAR). The population structure was analyzed with the “Structure 2.3.1” freeware with admixture (7). The “Estimated Ln Prob of Data” obtained from running the software was considered to estimate the number of subpopulations (K). This data corresponds to the estimation of ln Pr(X/K), where X are the genotypes. To verify the reliability of the estimations obtained by running the software, three runs were performed for each K.
Results
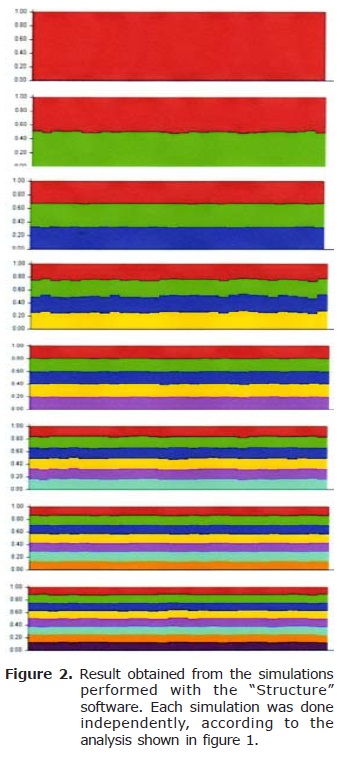
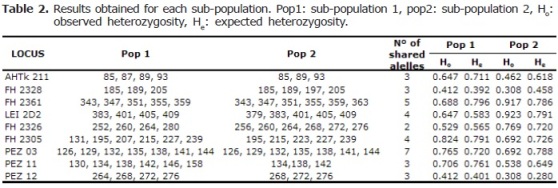
The results obtained from the simulations performed with the “Structure 2.3.1” software are shown in figures 1 and 2. Obtained alleles, observed and expected heterozygosity for each population are shown in table 2.
The number of subpopulations (K), according to the Bayesian rule [considering K=1, where the highest value of ln Pr(X/K) was obtained] was inferred according to Pritchard et al (5).
Discussion
According to the data obtained with the software “Structure”, it is not possible to establish a subdivision between the animals of the sample, as all of them belong to the same population. The analysis of the population presented in this paper show that the method has power to detect how many populations are presented in the samples studied.
The “Structure” software has been successfully used in different species, with the aim of identifying subpopulations of animals of different breeds or areas (8,9).
The analysis of our populations of “Cimarron Uruguayo” dogs, performed with the “Structure” software, gave a value of K=1 (Figures 1 and 2.). This would imply that the analyzed animals belong to the same population. Previous studies performed with RAPDs showed a high homogeneity and genetic identity among the animals of the NE and S areas (10), which would match the results obtained in this work. However, when making this type of studies, microsatellites have some advantages, including their high variability in populations and their codominant inheritance, which have made them more widely used (11).
In dogs, particularly, this software has been used with the aim of identifying subpopulations corresponding to different breeds (9). In our case, the result coincides with the history of the dog breed “Cimarron Uruguayo”, as it would confirm the hypothesis that current dogs derive from the founding nucleus that managed to take refuge in the NE region of the country (12). In addition, the distribution of the breed among the different areas of Uruguay continues nowadays, so there is no isolation among the different groups of animals, and the exchange is constant. This would not only not allow the formation of separated subpopulations, but also all the animals would continue being part of the same population. However, we intend to continue with these kinds of studies with other methods as population and quantitative genetics.
Acknowledgments
The authors would like to thank the Uruguayan Kennel Club for their collaboration in the sample extraction and the CSIC for their economical support.
References
1. Kennel Clud Uruguayo (KCU). Standard del perro Cimarrón Uruguayo. [En linea]. [Consultado Agosto de 2005]. URL Disponible en: http://www.kcu.com.uy/cimarron_std.htm. [ Links ]
2. Assunçao F. El Perro Cimarrón. 1ª ed. Montevideo: Linardi y Risso. 2008. [ Links ]
3. Gagliardi R, Martínez M, Llambí S, García CB, Arruga MV. El perro Cimarrón Uruguayo. Espécies 2008; 113:32-33. [ Links ]
4. Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MAM, et al. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 2001; 159:699-713. [ Links ]
5. Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000; 155: 945-959. [ Links ]
6. Gagliardi R. Estudios genéticos en caninos de raza Cimarrón Uruguayo (Canis familiaris). [Tesis de Maestría]. Uruguay: Universidad de la República, PEDECIBA; 2009. [ Links ]
7. Jonathan K. Pritchard, Xiaoquan Wen, Daniel Falush. Documentation for structure software: Version 2.2. Chicago: Department of Human Genetics. University of Chicago. [En linea]. URL Disponible en: http://pritch.bsd.uchicago.edu/structure.html [ Links ]
8. Padilla JA, Calero R, Sansinforiano ME, Parejo JC, Martínez-Trancón M, et al. Bayesiana de Estructura Genética Poblacional en la Raza Bovina Blanca Cacereña. XII Jornadas de Producción Animal. 2007. 16-17 de mayo. Zaragoza, España: Asociación Interprofesional para el Desarrollo Agrario; 2007. [ Links ]
9. Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, et al. Genetic Structure of the Purebred Domestic Dog. Science 2004; 304:1160-1164. [ Links ]
10. Llambí S, Gagliardi R, Martínez M, Estevez J, Gorozurreta A, Costa G, et al. Analysis of two populations of the Uruguayan canine breed Cimarron (Canis familiaris) using rapd markers. Rev MVZ Cordoba 2008; 13:1464-1468. [ Links ]
11. Morera Sanz L, de Andrés D, Barbancho J, Garrido J, Barba C. Detección de Variabilidad Genética por Microsatélites en el Alano Español. Archivos de Zootecnia 1999; 48: 72-77. [ Links ]
12. Silveira C, Fernández G, Barba C. El Perro Cimarrón, la Raza Canina Autóctona del Uruguay. Archivos de Zootecnia 1998; 47: 533-536. [ Links ]