Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista MVZ Córdoba
Print version ISSN 0122-0268
Rev.MVZ Cordoba vol.19 no.1 Córdoba Jan./Apr. 2014
ORIGINAL
Relationship between apoptosis and the BH2 domain sequence of the VP5 peptide of infectious pancreatic necrosis virus
Relación entre apoptosis y secuencia del dominio BH2 del peptido VP5 del virus de la necrosis pancreatica infecciosa
Cesar Ortega S,1* Ph.D, Sylvia Rodríguez S,2 Ph.D, Juan Carlos Espinoza,3 M.Sc, Juan Kuznar,3 Ph.D, Alex Romero,4,5 Ph.D, Ricardo Enríquez,4 Ph.D.
1Universidad Autonoma del Estado de México, Faculty of Veterinary Medicine and Animal Husbandry, Advanced Animal Health Research and Study Center. Toluca, Mexico, AP. 4-56.
2Biologic Research Center (High Council for Scientific Research) c/ Ramiro de Maeztu 9 Madrid 28040, Spain.
3Universidad de Valparaiso, Faculty of Sciences, Biochemistry and Virology Laboratory. Gran Bretaña 1111, Valparaíso, Chile.
4Universidad Austral de Chile, Faculty of Veterinary Sciences, Biotechnology and Aquatic Pathology Laboratory. Valdivia, Chile.
5Centro FONDAP: Interdisciplinary Center for Aquaculture Research (INCAR), Chile.
*Correspondence: cos_mx@hotmail.com
Received: March 2013; Accepted: September 2013.
ABSTRACT
Objective. To determine whether the level of apoptosis induced by infectious pancreatic necrosis virus (IPNV) is related to the amino acid sequence of the BH2 domain of the VP5 protein and the level of infectivity. Materials and methods. Three IPNV strains were used, the VP2 protein gene was amplified for genotyping and the VP5 sequence was also obtained. The infectivity of the strains was calculated using the viral titer obtained at 12, 24, 36 and 45 hpi in CHSE-214 cells. The percentage of apoptosis in infected cells was visualized by TUNEL assay and immunohistochemistry (caspase 3 detection). Results. The V70/06 and V33/98 strains corresponded to genotype Sp, while V112/06 to VR-299; the amino acid analysis of the V70/06 strain allows its classification as middle virulent strain and V33/98 and V112/06 strains as low virulent ones; infection with the V112/06 strain produced a lower viral titer (p<0.05). The VP5 gene of the 3 strains showed four homologous domains to Bcl-2, however, the BH2 domain was truncated in V70/06 and V33/98 (12 kDa), being complete (15kDa) in V112/06, which also showed the Trp155 residue, equivalent to Trp188 considered as a critical factor for the function of Bcl-2. The average apoptosis was below 12%, showing no differences between strains (p>0.05). Conclusions. The results showed that the differences in the BH2 sequence of the VP5 protein, infectivity and the VP2 sequence are not associated with the modulation of apoptosis.
Key words: Apoptosis, Infectivity, IPNV, VP5 (Sourse: MeSH).RESUMEN
Objetivo. Determinar si el nivel de apoptosis inducido por cepas del virus de la necrosis pancreática infecciosa (IPNV) tiene relación con la secuencia aminoacídica del dominio BH2 de la proteína VP5 y el nivel de infectividad. Materiales y métodos. Se utilizaron tres cepas de IPNV; el gen de la proteína VP2 fue amplificado para genotipificación y se obtuvo la secuencia de VP5. La infectividad de las cepas se calculó mediante el título viral obtenido a 12, 24, 36 y 45 hpi en células CHSE-214. Los porcentajes de apoptosis en células infectadas se visualizaron mediante ensayo TUNEL e inmuno-histoquímica (detección de caspasa 3). Resultados. Las cepas V70/06 y V33/98 correspondieron a genotipo Sp, mientras que V112/06 a VR-299; el análisis aminoacídico relacionó a V70/06 como cepa de mediana virulencia y a V33/98 y V112/06 de baja virulencia; la infección con V112/06 produjo menor título viral (p<0.05). El gen VP5 de las 3 cepas presentó los cuatro dominios homólogos a Bcl-2; sin embargo, el dominio BH2 fue truncado en V70/06 y V33/98 (12 kDa); siendo completo (15kDa) en V112/06, que además, presentó el residuo Trp155, equivalente a Trp188 considerado factor crítico para la función de Bcl-2. El promedio de apoptosis fue inferior a 12%, no se observaron diferencias entre cepas (p>0.05). Conclusiones. Los resultados mostraron que las diferencias en la secuencia de BH2 de la proteína VP5, la infectividad y en la secuencia de la proteína VP2 no están asociadas con la modulación de apoptosis.
Palabras clave: Apoptosis, infectividad, IPNV, VP5 (Fuente: MeSH).
INTRODUCTION
The infectious pancreatic necrosis virus (IPNV) causes a contagious globally distributed disease that mainly affects young salmonids (1); the species, age and health condition of animals affected and the virulence of the strain are factors that influence the clinical signs and mortality level (2,3).
The IPNV genome consists of two double-stranded RNA segments (dsRNA). Segment A exhibits two open reading frames (ORFs); the highest ORF encodes a polyprotein 5'-pVP2-NS-VP3', which is cotranslationally divided by the nonstructural protease (NS) or VP4, generating structural proteins VP2 and VP3, which respectively comprise the external and internal surface of the viral capsid; the lower ORF encodes an NS peptide known as VP5 which is not part of the virion and has been implicated in the inhibition of apoptosis, thus promoting viral survival. The Segment B shows a single ORF encoding a polymerase RNA dependent on RNA (RdRp), known as VP1 (3,4).
The role of IPNV proteins as virulence factor is not fully determined, but it is suggested that through any of its products, the virus could modulate certain cellular processes to favor the survival of its progeny, in addition, to alter defense mechanisms for establishing the infection (5,6). Sano et al (7), related the virulence of IPNV with products of the genome of segment A, and subsequently, Song et al (4) identified that variations in residues 217 and 221 in the amino acid sequence of the VP2 protein influence the virulence level of strains. The function of VP5 is uncertain, but since it shows four homologous domains with anti-apoptotic cellular proteins Bcl-2 (BH) it is considered as virulence factor that could modulate this form of cell death (6,8,9), associated with the amino acid sequence of the BH2 domain (10).
Apoptosis is a genetically controlled cell death process, involved in regulating the development and homeostasis of tissues; it is also a defense mechanism that can be stimulated by immune reactions or when there is cell damage by disease or toxic agents. This process can be initiated by external (extrinsic or receptor-mediated) or internal (intrinsic or mitochondrial) cell stimulation; both involve a preserved group of specific aspartate cysteine-proteases called caspases that are synthesized as zymogens, which must undergo proteolytic fragmentation to activate. The activation of the initial caspase triggers a sequential death program, where executing caspases alter proteins of the cytoskeleton and nuclear matrix. The activation of caspase 3 releases the inhibitor of a cytoplasmic DNase (CAD) that gives rise to the internucleosomal fragmentation of DNA (9,11,12). In viral infections, the modulation of apoptosis has been associated with products of the virus, which can manipulate certain cells processes to allow the survival of their progeny (13-15).
Hong et al (5) reported that the cytopathic effect (CPE) showed by cells infected with IPNV is a post apoptotic process, related to the suppression of the gene of the survival factor Mcl-1, a member of the antiapoptotic protein family Bcl-2; in contrast, Espinoza et al (13), reported that only 12% of the cell population infected with a strain of serotype VR-299 showed apoptosis, with cell death by necrosis taking precedence. Santi et al (10), observed no differences in the percentage of apoptosis in cells infected with recombinant strains of Spjarup serotype (Sp), including a strain that did not express VP5, suggesting that replacements or absences of certain residues of the sequence of the BH2 domain or other factor(s) of the virus could influence the development of this form of cell death. With this background, this study was undertaken to relate the induction level of apoptosis with the BH2 domain sequence and with the level of infectivity of three field strains of IPNV.
MATERIALS AND METHODS
Cells and virus. Chinook salmon embryo (CHSE-214) cells were used for the propagation of IPNV and in assays to assess apoptosis. Cells were cultured at 20°C in a Minimal Essential Medium with Eagle (E-MEM) salts, supplemented with 100 IU of penicillin/ml, 100 µg of streptomycin/ml and 10% (v/v) of fetal bovine serum (FBS). Once the monolayer has been formed, cells were kept in E-MEM supplemented with 2% (v/v) of FBS.
The virus strains used were obtained from clinical cases of infectious pancreatic necrosis (IPN). The V33/98 virus was isolated in 1998 from juvenils of Atlantic salmon (Salmo salar) the infection of which caused a mortality of 80%; the V70/06 virus was obtained in 2006 from juvenils of S. salar in which high initial mortality was observed with evolution to low mortality and chronicity, while the V112/06 virus was isolated from rainbow trout fries (Oncorhynchus mykiss) where a mortality of 40% was observed (16). The virus was reactivated by inoculation in pre-confluent CHSE-214 cells in E-MEM with 2% (v/v) of FBS; when showing an extensive cytopathic effect (CPE), the culture was frozen for 24 h and then thawed and centrifuged for 10 min at 4,000 rpm at 4°C; the viral titer was calculated by the Reed-Muench method (17).
The following access codes to the GenBank were used for the alignment and comparison of the VP5 protein of IPNV: AAK71697 VP5 Ab/E1S; AY379740 VP5 of 12.1 kDa/Sp; Q69CI1 VP5 15 Kda IPNV/Sp; AAA92630 VP5 IPNV/VR-299; P10415 Bcl2 Humana-Apoptosis regulator.
Amplification of VP2 y VP5 genomes by RT-PCR. The viral RNA was isolated from cell culture supernatants by column extraction according to specifications of the Viral RNA mini Kit Q1Aamp® (Cat. No. 52904; QiAGEN Germany). The RNA diluted in 50 µl of sterile deionized water treated with diethyl pyrocarbonate was quantified by spectrophotometry and frozen at -80°C until use.
The obtaining and amplification of cDNA was carried out using 0.25 µg of Random primers (Promega) in 20 µl of reagent solution. The amplification of a gene product of 1180 pb corresponding to a portion of the VP2 coding sequence was carried out in a thermal cycler Gene Amp PCR System 2400® (Perkin Elmer) in a volume of 50 µl; the primers used for the amplification of VP2 and cycle parameters were the same as described in Ortega et al (16). The amplification of the VP5 gene was carried out with quantities of reagents and similar cycles as for VP2, replacing specific primers to amplify a product of 450 pb: Fwd-5'ATGGCGAAGCCCTTTCTAAC-3' and Rev-5'ACAGACTTCCTTCGAAGGTG-3'; both products were subjected to electrophoresis in agarose gel at 1% with ethidium bromide, applying a current of 80 mA for 30 min in a camera SUB-CELL GT® and power supply POWER-Cap 200® (BIO-RAD). The gels were analyzed in a transilluminator TFX-20.M® (VILBER LOURMAT), detecting fragments of 1.180 bp and 450 bp, corresponding to VP2 and VP5, respectively.
Segments of cDNA with a portion of the VP2 and VP5 coding sequence were extracted from the agarose gel according to the instructions of the supplier of the E.Z.N.A.® Gel Extraction Kit (OMEGA BIO-TEK) and once purified, were sent to be sequenced. The nucleotide sequences obtained were translated using ExPASy and the amino acid sequence deduced of both proteins was compared with sequences obtained from NCBI, using the Molecular Evolutionary Genetics Analysis software version 4.0 MEGA4 (4,16).
Determination of the viral titer in cell culture supernatant. For determining differences in the level of infectivity and replication of the viruses used, CHSE-214 cells were inoculated to a infection multiplicity MDI=0.1 in 25 cm2 culture bottles and after 12, 24, 36 and 45 h of incubation at 15°C, the bottles were frozen and thawed twice; the content was centrifuged at 4.000 rpm for 10 min at 4°C and the supernatants were tittered in accordance with the Reed and Muench method (17). The titers obtained were represented by the average of three samples with a standard deviation indicator bar.
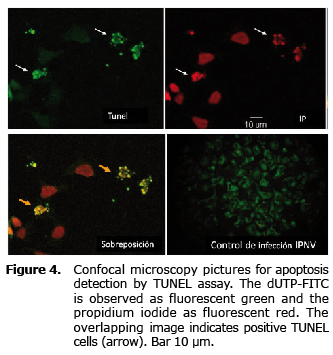
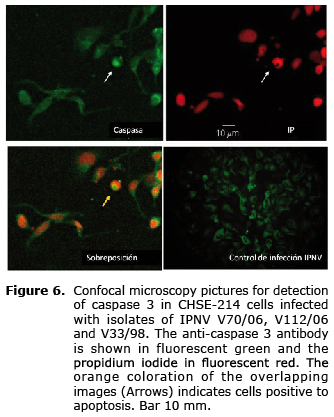
Detection of apoptosis induction in CHSE-214 cells. CHSE-214 cells with a confluence of 90% in circular glass coverslips were infected by triplicate at MDI=10 with each of the strains; three coverslips were not infected to be used as negative control. After 8, 12 and 18 h of incubation at 15°C, the samples were washed in PBS pH 7.2 and fixed for 15 min in paraformaldehyde at 4%. Apoptosis was detected by the identification of the DNA fragmentation by TUNEL assay (Terminal Transferase Uridyl Nick End Labeling) according to instructions from the supplier Apoptosis Detection Kit (USBiological T9162-180), and also by detection of caspase 3 by immuno-histochemical assay, following the recommendations of the manufacturer of the product Anti-ACTIVE® Caspase-3 pAb. Samples were analyzed under a confocal microscope; the number of positive cells in a sample of approximately 300 cells was obtained, and was represented as the average of three independent samples ± standard error of the mean (SEM). Data were analyzed by the Friedman test, with statistical significance defined by p<0.05 values.
RESULTS
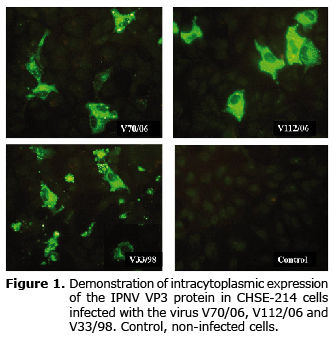
Thirty-six hours after the infection (hpi), the three virus strains caused multiple cytopathic effect foci (CPE) in infected cells; the presence and intracytoplasmic distribution of the viral antigen (VP3 protein) was diffusely detected by indirect immunofluorescence (IFAT) showing areas of aggregation or inclusion bodies. The detection of the antigen occurred later in V112/06. Non-infected cells used as negative control showed no fluorescence (Figure 1).
The gene fragments of 1180 and 450 pb, corresponding to the VP2 and VP5 proteins of IPNV, were detected by RT-PCR in cells infected with the three strains (not shown). The amino acid sequence of VP2 was compared with the sequence of the Sp (Genbank AJ829474) and VR-299 strains (L40584); the sequence of the V70/06 (Genbank GU072916) and V33/98 strains (Genbank GU072915) corresponded to genogroup Sp, which show identity percentages of 98 and 96%, respectively; the sequence of the V112/06 strain (Genbank GU072914) showed a identity percentage of 96% with genogroup VR-299.
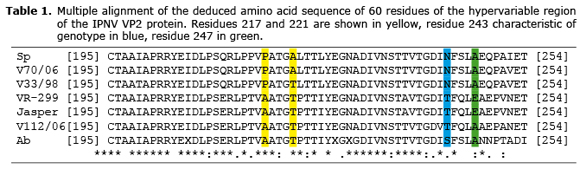
The analysis of the VP2 sequence shows that the residues involved in the virulence or infectivity of the IPNV in the V70/06 strain corresponded to Pro-217 and Ala-221; Ala-217 and Thr-221 for V112/06, and finally Pro-217 and Thr-221 for V33/98 (Table 1).
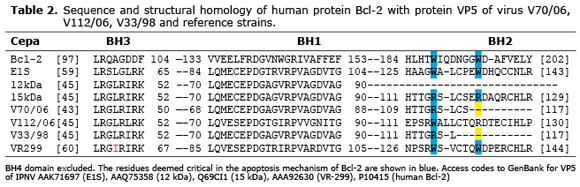
Regarding VP5, the genes of the three strains showed the four homology domains with Bcl-2, and initiated translation at codon 16. However, the VP5 ORF in V112/06 was 394 pb, coding 131 aa (15 kDa); while in V70/06 and V33/98 the ORF was composed of 351 pb, coding a truncated protein of 116 aa (13 kDa).
The multiple alignment of VP5 sequences of V70/06 (JQ247979), V112/06 (JQ253903) and V33/98 (JQ253902), with the partial sequence of human protein Bcl-2 (P10415) and VP5 of strains E1S, 12 kDa and 15 kDa (10), was similar in BH4, BH1, and BH3 domains. However, the BH2 domain was only complete in V112/06, and partially truncated in V70/06 and V33/98. Residues 115 and 122 of VP5, which are homologous to residues Trp188 and Trp195 of Bcl-2 and that are considered critical to the antiapoptotic function of this protein, were Trp115 and Arg122 for V112/06; while V70/06 and V33/98 only showed Arg115 (Table 2).
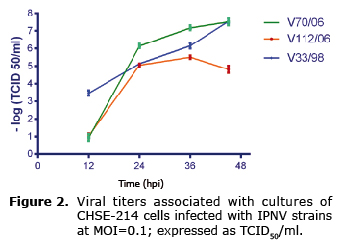
In general, the viral titers of the three strains were increased as the infection spread. At 12 hpi, cells infected with V70/06 and V112/06 showed a viral titer ≤ 1x100.66 TCID50/mL, while in those infected with V33/98 it was 1x103.5. At 24 and 36 hpi, V70/06 showed more titers than the other two strains, but at 45 hpi its titer was similar to V33/98 (1x107.5). The titers in V112/06 were lower at 36 and 45 hpi (Figure 2).
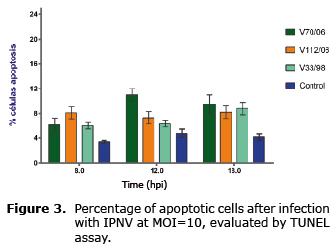
The percentage of apoptosis obtained in cells infected with the three strains was similar (p>0.05); there was no gradual increase between the times analyzed, and in any case the percentage was not greater than 12%. In cells infected with V70/06, the percentage of apoptosis increased between 8 and 12 hpi, without further variation (Figure 3). Cells used as negative control and not infected showed a lower percentage of apoptosis with respect to infected samples.
Figure 4 shows positive samples to the reaction of the TUNEL assay and positive cells to the IPNV infection.
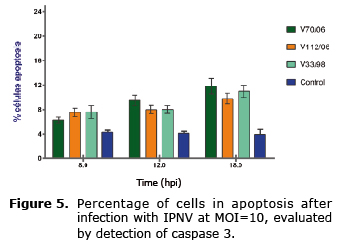
The activation profile of caspase 3 examined by immunoassay (Figure 5) was similar among the three strains; despite the fact that at 18 hpi it is possible to observe a slight increase with respect to the previous reading, there were no differences between evaluated times (Figure 6), and the percentage of cells in apoptosis did not exceed 12% in the analyzed times (p>0.05).
DISCUSSION
Several products of RNA virus genes affecting fish have been reported as regulators of apoptosis (14); since that VP5 protein of IPNV has four homology domains with proteins of the subfamily Bcl-2 (BH) with antiapoptotic activity, it has been suggested that during replication the virus could inhibit the apoptosis to permit the viral replication.
In this study, the VP5 protein of the strains used showed the four BH domains characteristic of Bcl-2 antiapoptotic proteins. However, the BH2 domain, which is considered critical to the antiapoptotic activity, was truncated at V70/06 and V33/98, and was complete in the V112/06 strain; despite this, there were no differences in the induction level of apoptosis between strains, and the average did not exceed 12% in the times evaluated. This indicates that the size of VP5 has no influence on the induction of apoptosis and is consistent with what was reported by Santi et al (10), who observed that IPNV may induce apoptosis regardless of the size and expression of the VP5 protein. A similar situation has been reported with the virus of the African swine fever that contains a homologous protein to the inhibitor of apoptosis of baculovirus (IAP) that is not necessary for in vitro viral growth, does not modulates virulence and has no influence on the induction of apoptosis.
As a possible explanation for the above, Cuconati and White (18), pointed out that certain viral Bcl-2 proteins may have antiapoptotic activity even lacking some of the BH domains, but that the BH2 domain is a key element for heterodimerization with the BAX and BAK death promoters. In the case of IPNV, this interaction would take place through the residue Trp155 of VP5 that would correspond to residue Trp188 of Bcl-2, considered a critical residue for fulfilling the antiapoptotic function; accordingly, strains that express Trp-155 in VP5 should inhibit apoptosis. In this regard, Santi et al (10) proposed that the fact of not observing differences in the induction level among IPNV strains used in her study, could be due to the absence of the Trp155 residue. However, although in the present study the VP5 of V112/06 showed the Trp-115 residue in BH2, there was no difference in terms of induction with respect to other strains, so the sequence or substitution of this residue is not a critical factor in the modulation of apoptosis.
The results obtained in this study showed that the IPNV VP5 protein would not participate in the modulation of apoptosis, contrary to what was observed in the infection with the infectious bursa disease virus (IBDV), that encodes a homologous protein containing a transmembrane region, but lacks homology domains with Bcl-2, shows a PEST domain that is usually present in short-lived and early-expression proteins that fulfill regulatory activities (19). During the replication of IBDV, VP5 accumulates in the cell membrane, alters its morphology and causes subsequent rupture, acting as death factor allowing the release of its progeny (12, 20, 21).
The involvement of the VP2 protein in the virulence or infectivity of IPNV is associated with its amino acid sequence; strains that show Thr-217 and Ala-247 residues produce more viral titer during a replication cycle, and are considered of greater virulence (4). In this study, it was observed that the infectivity of the strains used was unrelated to the induction level of apoptosis in CHSE-214 cells; strains V70/06 and V33/98 corresponded to average virulence strains by having the residues Pro-217 and Ala-247, and Pro-217 and Thr221 respectively, while in V112/06 sequence was Ala-217 and Ala-247, related to avirulent strains (4). While overall the replication curves of the three strains showed a similar growth pattern, V112/06 produced less titers, confirming it as a strain with lower infectivity.
The percentage of apoptosis obtained through TUNEL and immunoreaction assay does not show differences in cells infected with the strains V70/06 and V33/98 of the Sp genotype and strain V112/06 of genotype VR-299, evidencing that the genotype of the strain is not related to this form of cell death. It has been indicated that the IPNV dsRNA genome can trigger apoptosis by the activation of the PKR pathway that inhibits the translation of cellular proteins, and the RNase L pathway that degrades the mRNA (22-24); more recently, Chiu et al (25), observed that the VP3 protein induces the expression of BAD stimulating apoptosis through the mitochondrial pathway. These assertions would support the fact that apoptosis observed during infection by IPNV may develop for independent reasons from the characteristics of the strain involved in the infection (14, 26). In support of the foregoing, Imajoh et al (14) reports that Salmo salar cells positive to IPNV infection do not frequently express this form of death; suggesting that apoptosis corresponds to an autonomous suicide process for eliminating infected cells preventing the viral spread.
After infecting CHSE-214 cells with Ab serotype IPNV, considered of low virulence with respect to Sp and VR-299 strains, Hong and Wu (27) determined that apoptosis is related with the expression of the BAD death gene, as an early stress factor induced through TNF receptors. In relation to this, Espinoza et al (13) propose that if apoptosis is initiated via the receptor, the viral serotype does not affect the activation of the process. This proposal is consistent with that observed in this study, where there are no differences in the average of apoptosis obtained in cells infected with V70/06 and V33/98 Sp serotype and V112/06 VR-299 serotype strains. However, the receptor that could modulate the process is unknown.
Taking into account that apoptosis is an early defense mechanism, the level obtained with the three strains of this study evidences that IPNV is a poor inducer of this death mechanism, which is consistent with Espinoza et al (13), who observed that the percentage of apoptotic cells was less than 12%, while at 15 hpi 75% of the cell population showed necrosis; the results are also consistent with that reported by Hong and Wu (27), who observed that although practically all cells were positive to the infection, only a low percentage developed apoptosis.
In this study it was observed that the percentage of apoptosis during infection was unrelated to the presence of the Trp-155 residue in the sequence of the IPNV BH2 domain; this result is consistent with researches indicating that apoptosis occurs irrespective of the presence or sequence of certain viral peptides with Bcl-2 homology (14, 18, 26). Likewise, since neither the differences in the VP2 and VP5 sequence nor the length of the latter influence the degree of apoptosis, probably other variables such as the type of tissue and host susceptibility would have greater relevance, as observed in the case of ISAV (28). However, the actual participation of the viral components in the induction of this form of cell death and pathogenesis is yet to be determined.
In conclusion, differences in the BH2 sequence of the VP5 protein and the infectivity in the sequence of the VP2 protein are not associated with the modulation of apoptosis.
Acknowledgements
To The National Science and Technology Council (Conacyt) project 99736 and project PROMEP 103.5/09/4196; to the Graduate School and the Biotechnology and Aquatic Pathology Laboratory of the Animal Pathology Institute, Faculty of Veterinary Sciences of Universidad Austral de Chile.
REFERENCES
1. Roberts R, Pearson M. Infectious Pancreatic Necrosis in Atlantic salmon, Salmo salar L. J Fish Dis 2005; 28:383-389. [ Links ]
2. Bruslind L, Reno P. Virulence Comparison of Three Buhl-Subtipe Isolates of Infectious Pancreatic Necrosis Virus in Brook Trout Fry. JAAH 2000; 12:301-315. [ Links ]
3. Shivappa R, Song H, Yao K, Aas-Eng A, Evensen Ø, Vakharia V. Molecular characterization of Sp serotype strains of infectious pancreatic necrosis virus exhibiting differences in virulence. Dis Aquat Org 2004; 61:23-32. [ Links ]
4. Song H, Santi N, Evensen Ø Vakharia V. Molecular Determinants of Infectious Pancreatic Necrosis Virus Virulence and Cell Culture Adaptation. J Virol 2005; 79:10289-10299. [ Links ]
5. Hong J, Hsu Y, Wu J. Infectious pancreatic necrosis virus induces apoptósis due to down-regulation of survival factor MCL-1 protein expression in a fish cell line. Virus Res 1999; 63:75-83. [ Links ]
6. Santi N, Song H, Vakharia V, Evensen Ø. Infectious Pancreatic Necrosis Virus VP5 Is Dispensable for Virulence and Persistence. J Virol 2005; 79:9206-9216. [ Links ]
7. Sano M, Okamoto H, Fukuda H, Saneyoshi M, Sano T. Virulence of infectious pancreatic necrosis virus is associated with the larger RNA segment (RNA segment A). J Fish Dis 1992; 15:283-293. [ Links ]
8. Hong J, Gong H, Wu J. IPNV VP5, a Novel Anti-apoptósis gene of the Bcl-2 Family, Regulates Mcl-1 and Viral Protein Expression. Virol 2002; 295:217-229. [ Links ]
9. Weber S, Fichtner D, Mettenleiter T, Mundt E. Expression of VP5 of infectious pancreatic necrosis virus strain VR299 is initiated at the second in-frame start codon. J Gen Virol 2001; 82:805-812. [ Links ]
10. Santi N, Sandtrø A, Sindre H, Song H, Hong J, Thu B, Wu J, Vakharia V, Evensen Ø. Infectious Pancreatic Necrosis Virus induces apoptósis in vitro and in vivo independent of VP5 expression. Virol 2005; 342:13-25. [ Links ]
11. Hong J, Lin T, Hsu Y, Wu J. Apoptósis Precedes Necrosis of Fish Cell Line with Infectious Pancreatic Necrosis Virus Infection. Virol 1998; 250:76-84. [ Links ]
12. Galloux M, Libersou S, Morellet N, Bouaziz S, Da Costa B, Ouldali M, Lepault J, Delmas B. Infectious Bursal Disease Virus, a Non-enveloped Virus, Possesses a Capsid-associated peptide That Deforms and Perforates Biological Membranes. J Biol Chem 2007; 282:20774-20784. [ Links ]
13. Espinoza J, Cortés M, Kuznar J. Necrosis of infectious pancreatic necrosis virus (IPNV) infected cells rarely is preceded by apoptósis. Virus Res 2005; 109:133-138. [ Links ]
14. Imajoh M, Hirayama T, Oshima S. Frequent occurrence of apoptósis is not associated with pathogenic Infectious Pancreatic Necrosis Virus (IPNV) during persistent infection. Fish Shellfish Immunol 2005; 18:163-177. [ Links ]
15. O'Brien V. Viruses and Apoptósis. J Gen Virol 1998; 79:1833-1845. [ Links ]
16. Ortega C, Rodríguez S, de las Heras A, Romero A, Monrás M, Enríquez R. Evaluation of the level of Mx3 protein synthesis induced by infectious pancreatic necrosis virus (IPNV) strains of different infectivity. Vet Immunol Immunopathol 2011; 141:190-200. [ Links ]
17. Reed J, Muench H. A simple method for estimating fifty percent end points. Am J Hyg 1938; 27:493-497. [ Links ]
18. Cuconati A, White E. Viral homologs of BCL-2: role of apoptósis in the regulation of virus infection. Genes & Dev 2002; 16:465-2478. [ Links ]
19. Liu M, Vakharia V. Nonstructural Protein of Infectious Bursal Disease Virus Inhibits Apoptósis at the Early Stage of Virus Infection. J Virol 2006; 80:3369-3377. [ Links ]
20. Lombardo E, Maraver A, Espinosa I, Fernández-Arias A, Rodríguez J. VP5, the Nonstructural Polypeptide of Infectious Bursal Disease Virus, Accumulates within the Host Plasma Membrane and Induces Cell Lysis. Virol 2000; 277:345-357. [ Links ]
21. Brandt M, Yao K, Liu M, Heckert R, Vakharia V. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J Virol 2001; 75:11974-11982. [ Links ]
22. Wolf K. Fish viruses and fish viral diseases. Ithaca, N.Y. Canstock Publishing Associates-Cornell University Press, 1988. [ Links ]
23. Lyles D. Cytopathogenesis and Inhibition of Host Gene Expression by RNA Viruses. Microbiol Mol Biol Rev 2000; 64:709-724. [ Links ]
24. Robertsen B. The interferon system of teleost fish. Fish Shellfish Immunol 2006; 20:172-191. [ Links ]
25. Chiu CL, Chou YL, Wu JL, Hong JR. Aquatic birnavirus capsid protein, VP3, induces apoptosis via the Bad-mediated mitochondria pathway in fish and mouse cells. Apoptosis 2010; 15:653-668. [ Links ]
26. Eléouet J, Druesne N, Chilmonczyk S, Momge D, Dorson M, Delmas B. Comparative Study of In-situ Cell Death Inducid by the Viruses of Viral Haemorrhagic Septicaemia (VHS) and Infectious Pancreatic Necrosis (IPN) in rainbow trout. J Comp Pathol 2001; 124:300-307. [ Links ]
27. Hong J, Wu J. Induction of apoptotic death in cells via Bad gene expression by Infectious pancreatic necrosis virus infection. Cell. Death Differ 2002; 9:113-124. [ Links ]
28. Joseph T, Cepica A, Brown L, Ikede B, Kibenge F. Mechanism of cell death during infectious salmon anemia virus infection is cell type-specific. J Gen Virol 2004; 85:3027-3036. [ Links ]