Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista MVZ Córdoba
Print version ISSN 0122-0268
Rev.MVZ Cordoba vol.19 no.2 Córdoba May/Aug. 2014
ORIGINAL
Association of gene BoLA DRB3.2 with production traits in a dairy herd of Antioquia, Colombia
Asociación del gen BoLA DRB3.2 con características productivas en un hato lechero de Antioquia, Colombiaá
Juan Zambrano A,1 M.Sc, Julián Echeverri Z,2 Ph.D, Albeiro López-Herrera,2* Ph.D.
1Universidad Nacional de Colombia sede Medellín, Facultad de Ciencias, Posgrado en Biotecnología, Grupo de Investigación BIOGEM. Medellí, Colombia.
2Universidad Nacional de Colombia Sede Medellín, Facultad de Ciencias Agropecuarias, Departamento de Producción Animal, grupo de Investigación BIOGEM, Calle 59A No 63-20 -Núcleo el volador, Bloque 50, Oficina 310, Medellín, Colombia.
*Correspondence: alherrera@unalmed.edu.co
Received: January 2013; Accepted: December 2013.
ABSTRACT
Objective. To determine the associations of BoLA DRB3.2 alleles present in Holstein and BON x Holstein cattle to production and milk quality traits in a dairy herd of Antioquia, Colombia. Materials and methods. Ninety-one cows, 66 Holstein and 25 BxH, were genotyped for the BoLA DRB3.2 gene, through PCR-RFLP technique. Furthermore, the association of the alleles of the gene BoLA DRB3.2 with milk yield (PL305), fat yield (PG305), protein yield (PP305) fat percentage (PGRA) and protein percentage (PPRO) were determined, using a general linear model. Results. Twenty-seven BoLA DRB3.2 alleles were identified; the most frequent alleles in Holstein were: BoLA DRB3.2*23, 22, and 24 with frequencies of: 0.159, 0.129, and 0.106, respectively and the most frequent alleles in BxH were: BoLA DRB3.2*23, 24 and 20 with frequencies of: 0.20, 0.140, and 0.120, respectively. Associations of BoLA DRB3.2 alleles with production and milk quality traits were also determined. In Holstein cows the BoLA DRB3.2*36 allele was associated with low PL305 (p≤0.01), high PGRA in multiparous cows (p≤0.05) and high PG305 in primiparous cows (p≤0.01). The BoLA DRB3.2*33 allele was associated with increased in the PPRO in multiparous cows (p≤0.01). In BXH cows only the BoLA DRB3*19 allele was associated with high PGRA (p≤0.05). Conclusions. The gene BoLA DRB3.2 shows high polymorphism in both groups; Holstein and BxH and some of its allelic variants were associated with production and milk quality traits.
Key words: DNA, genetic markers, milk protein, PCR (Source:CAB, AIMS).
RESUMEN
Objetivo. Determinar la asociación de los alelos BoLA DRB3.2 presentes en vacas Holstein y BONxHolstein con características productivas y de calidad composicional de la leche en un hato lechero de Antioquia, Colombia. Materiales y métodos. Fueron genotipificados 91 animales; 66 Holstein y 25 BONxHolstein (BxH) para el gen BoLA DRB3.2, mediante la técnica PCR-RFLP. También, se determinó la asociación de los alelos del gen BoLA DRB3.2 con producción de leche (PL305), producción de grasa (PG305), producción de proteína (PP305), porcentaje de grasa (PGRA) y porcentaje de proteína (PPRO); usando un modelo lineal generalizado. Resultados. Fueron identificados 27 alelos BoLA DRB3.2; los más frecuentes en Holstein fueron: BoLA DRB3.2*23, 22 y 24 con frecuencias de 0.159, 0.129 y 0.106 respectivamente y los más frecuentes para BxH fueron: BoLA DRB3.2*23, 24, 20 con frecuencias de 0.20, 0.140 y 0.120 respectivamente. También fueron determinadas asociaciones de los alelos BoLA DRB3.2 con características productivas y de calidad composicional de la leche. En vacas Holstein el alelo BoLA DRB3.2*36 fue asociado con baja PL305 (p≤0.01), alto PGRA en vacas multíparas (p≤0.05) y alta PG305 en vacas primíparas (p≤0.01). El alelo BoLA DRB3.2*33 fue asociado con alto PPRO en vacas multíparas (p≤0.01). En vacas BxH sólo el alelo BoLA DRB3.2*19 estuvo asociado con aumento en el PGRA (p≤0.05). Conclusiones. El gen BoLA DRB3.2 presentó un alto polimorfismo en ambos grupos genéticos; Holstein y BxH y algunas de sus variantes alélicas fueron asociadas significativamente con características productivas y características de calidad composicional de la leche.
Palabras clave: DNA, marcadores genéticos, PCR, proteína de la leche (Fuente: CAB, AIMS).INTRODUCTION
Milk production in Colombia has increased in an accelerated and sustained manner. In the year 1990, raw milk production was 3917 million liters, by the year 2012, it increased by 40%; reaching a production of 6518 million liters (1,2). This growth over the last 20 years shows not just the growth in number of animals, but also the use of bulls selected for their high genetic value for dairy production.
However, the intense selection of productive characteristics has led to an increase in the appearance of infectious diseases, metabolic problems, and diminished fertility in cattle (3). The costs associated to these types of problems, are mainly represented by the decrease in milk production, veterinary costs, premature discard of animals, milk rejection due to antibiotic contamination, among others (4).
There have been few and isolated intents in Colombia, to know the genetic value (genetic evaluations) of dairy cows and Bulls, and they have been undertaken, mainly to estimate volume of milk (5). Cattle ranchers, on their part, have aimed their production mainly towards milk volume, rather than towards quality, regarding composition and sanitary condition. Thus, strategies are required to allow improvement of production, milk quality, and cattle health.
Genetic improvement assisted by molecular markers is one of the alternatives that allows for the study of genes associated to productivity, milk quality and also to resistance or vulnerability to infectious diseases. The major histocompatibility complex (MHC), which in bovines is called Bovine Leucocyte Antigen (BoLA), has been widely studied, and some molecular markers associated to production, compositional quality of milk and sanitary problems have been identified, such as gene BoLA DRB3 (6).
Gene BoLA DRB3, is usually expressed by cells in the immune system such as macrophages dendritic cells or B lymphocytes, which process the antigen and then present it to the helper T lymphocytes to unleash the immune response against infectious pathogens(7). Their elevated level of expression and high polymorphism makes them potential molecular markers for Molecular Marker-assisted selection procedures (MAS). More than 100 different alleles (8,9) have been identified from exon 2, and some of them have been associated to the occurrence of infectious diseases and milk, fat and protein production (6,10-13).
The purpose of this investigation was to determine the association of the alleles of BoLA DRB3.2 gene with milk production and compositional quality traits in Holstein and BON x Holstein cattle, at a dairy farm in Antioquia, Colombia.
MATERIALS AND METHODS
Population of the study. The investigation was undertaken at the Paysandú farm, which is property of the Universidad Nacional de Colombia, Medellin branch, located in the department of Antioquia, Township of Santa Elena, 16 km east of Medellín in an area of lower montane rain forest (bmh-MB) with a mean temperature of 14°C at 2500 meters above sea level. 91 bovine females were genotipified at this farm for gene BoLA DRB3.2, 66 of them were Holstein and 25 were from the BxH genetic group.
Data recollection. Data of milk yield per lactation was gathered from the historical production data of the Paysandú farm and adjusted to 305 days (PL305). Milk production was measures with proportional flow meters. The characteristics of compositional quality of the milk were: fat percentage (PGRA) and protein percentage (PPRO), they were determined at the quality control lab of COLANTA, using the MilkoScan FT120® device, based on the FTIR principle (Fourier Transfer Infrared Spectroscopy).
Cows with different number of lactations were taken for this study (between one and eleven). For PGRA and PPRO cows with less than four data per lactation were included. Fat yield (PG305) and protein yield (PP305) in kg/lactation adjusted to 305 days was calculated as of the percentage of these parameters (PPRO y PGRA) and the milk yield adjusted to 305 days (PL305).
DNA extraction. 5 ml of peripheric blood was drawn into vacuum tubes with EDTA as anticoagulant, from each one of the 91 individuals. They were stored at 4°C until processed. DNA was extracted through the “salting out” method.
Amplification of the BoLA DRB3.2. gene. Exon 2 of gene BoLA DRB3 was amplified by semi-nested PCR, described by Van Eijk et al (14), with a few modifications. The primers used were: HL030 (ATCCTCTCTCTGCAGCACATTTCC), HL031 (TTTAAATTCGCG CTCACCTCGCCGCT) and HL032 (TCGCCGCTGCACAGTGAAACTCTC). The first amplification cycle was done at a reaction volume of 25 µl containing: 0.4 mM of each dNTP, 0.5 µM primer HLO30 and HLO31, 2.5 µl 10X magnesium free thermophilic buffer [500 mM KCl, 100 mM Tris-HCl (pH 9.0), 1% (v/v) Triton X-100], 2.5 µl of 25 mM MgCl2, 12.8µl of ultra-pure water, 1U Taq DNA polymerase (Fermentas, California, U.S.A) and 50 to 100 ng of genomic DNA. The reaction mixture took place in a thermo cycler (Biometra, GÖttingen Germany with the following temperature program: Initial denaturation at 94°C for 240 s, followed by 10 denaturation cycles at 94°C for 60 s, annealing at 60°C for 120 s, extension at 72°C for 60 s and a final extension at 72°C for 300 s.
The second PCR cycle was done at a reaction volume of 60 µl containing: 0.4 mM of each dNTP, 0.6 µM of each primer HLO30 and HLO32, 6 µl of 10X magnesium-free thermophilic buffer, 6 µl of 25 mM MgCl2, 36.9 µl of ultra pure water, 1U of Taq ADN polymerase (Fermentas, California, U.S.A) and 2.5 µl of amplified DNA from the first PCR cycle. The amplification program was as follows: Initial denaturation at 94°C for 240 s, followed by 25 denaturation cycles at 94°C for 60 s, annealing at 67°C for 120 s, extension at 72°C for 60 s, followed by a final extension of 72°C for 300 s. As negative control, reactions were undertaken without DNA. The resulting product was fixed in agarose gels at 1.5% (p/v) and visualized under ultraviolet light with a trans illuminator. (Biometra, Göttingen Germany).
Digestion with restriction enzymes. The amplification product obtained during the second PCR cycle was digested with three restriction enzymes RsaI, BstYI (New England BioLabs®) and HaeIII (Fermentas, California U.S.A) and separation of fragments was done by electrophoresis in agarose gel. Identification of the alleles of BoLA DRB3.2 was done by combination of the different patterns of restriction obtained in the following order: RsaI, BstYI, HaeIII for each sample, according to the allele nomenclature reported by Van Eijk et al (14). The identified BoLA DRB3.2 alleles for PCR-RFLP, were confirmed by sequencing, for which our amplifiers in the second PCR cycle were purified with the QIAquick kit (QIAGEN) and sent to Macrogene Inc in Seoul Korea for sample sequencing. The sequences obtained were aligned and analyzed with the BioEdit Sequence Alignment Editor program.
Determination of allelic frequencies. The frequency of the different alleles was done by determining the proportion of each form of the gene between the total numbers of copies of the studied population. The homozygotes (two copies of the same allele) and heterozygotes (one copy of each allele) were identified and the frequency for each allele was calculated by counting the homozygotes and adding half of the heterozygotes, with the method described by Laird and Lange (15).
Statistical analysis. An allele substitution model was used to determine the association of each BoLA DRB3.2 allele with the productive and milk quality traits, using the BoLA DRB3.2*24 substitution allele, according to what was described by Sharif et al (10) and Rupp et al (11). The model was described as follows:
Yijkl = µ + GGi + NPj + ΣkbkBoLAijk + eijkl
Where: Yijkl corresponds to the phenotype trait (PL305, PP305, PG305, PGRA and PPRO), µ is the general mean, GG corresponds to the i effect of the genetic group (i=1, 2), being 1 the Holstein genetic group and 2 the BxH genetic group, NP is the fixed effect of the j number of calvings (j=1,2), being 1 the primiparous cows (of one and two calvings) and 2 multiparous cows (of three or more calvings), bk is the regression coefficient of the number of copies of the k allele BoLA, BoLAijk corresponds to the fixed effect of the number of copies (0,1 and 2) of the BoLA k (k = 1......15) allele present in cow ijk and eijkl is the experimental error.
Dunnett‘s comparison test was used to determine statistical significance between the means of the BoLA DRB3.2 alleles compared with the mean of the substitution allele (allele 24) for each productive trait. The comparison analysis in the Holstein breed was done by comparing means of each one of the groups: primiparous and multiparous, independently for all the evaluated traits. For BxH the analysis was done in all the cows, due to the low number of animals (n=25) for all the evaluated traits. Alleles with frequency ≤2% were grouped in one category named “others”. The model was undertaken with the GLM (General Linear Model) procedure of SAS V9.1 (16).
RESULTS
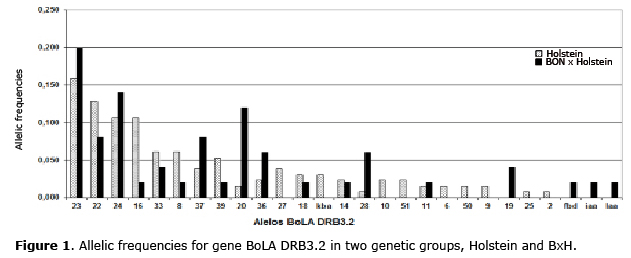
Allele Frequencies for gene BoLA DRB3.2. 23 alleles from the Holstein breed, and 18 alleles from the BxH genetic group were identified through PCR-RFLP 23 for a total of 27 allele variants of BoLA DRB3.2. The allelic frequencies for the Holstein group were located between 0.008 and 0.159 (light grey bars Figure 1), being the most frequent alleles 23, 22 and 24 with frequencies of 0.159, 0.129 y 0.106, respectively. In BxH the allele frequencies were between 0.02 and 0.20 (dark bars Figure 1), being 23, 24 and 20 the most frequent alleles with frequencies of 0.20, 0.14 and 0.12, respectively. It was determined that in the Holstein genetic group, 79% of the cows were heterozygote and 21% homozygote, for BxH a very similar result was obtained where 80% were heterozygote and 20% homozygote for gene BoLA DRB3.2.
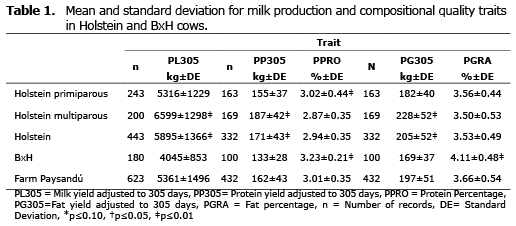
Milk Production. The average PL305 in the evaluated population was 5361 Kg (Table 1). The genetic group had a significant effect (p≤0.01), with a PL305 mean higher for Holstein (5895 kg), than for BxH (4045 kg). Likewise, the calving effect was significant (p≤0.01); primiparous Holstein cows presented a PL305 mean of 5316 kg, while the mean for multiparous was 6599 kg per lactation (Table 1).
Protein production and percentage. The mean for PP305 was 162 kg. The effect of the genetic group was significant (p≤0.01) for this characteristic, with a PP305 mean higher for Holstein (171 kg), than for BxH (133 kg). Likewise, the effect of the number of calvings was significant (p≤0.01); in the analysis done for Holstein cows; primiparous cows had a PP305 mean of 155 kg, while multiparous cows had a mean of 187 kg (Table 1). The mean for PPRO in the assessed population was 3.01% and the mean for each genetic group was significantly different (p≤0.01); for BxH it was higher with 3.23% and for Holstein it was 2.94%. Regarding the fixed effect of the number of calvings, it was significant for the Holstein breed (p≤0.01), where primiparous cows presented a PPRO mean of 3.02% and multiparous were 2.87% (Table 1).
Fat Production and Percentage. PG305 for the assessed population was 197 kg. The fixed effect of the genetic group was significant (p≤0.01), with a mean of PG305 higher for the Holstein breed (205 kg), than for BxH (169 kg). Likewise, the fixed effect of the number of calvings in Holstein cows was significant (p≤0.01); primiparous cows had a PG305 mean of 182 kg, while multiparous cows had a mean of 228 kg (Table 1). The mean for PGRA in the assessed population was 3.66% and the means for each genetic group were significantly different (p≤ 0.01); for BxH it was 4.11% and for Holstein it was 3.53% regarding calving effect, there were no significant differences found in Holstein, primiparous cows had a PPRO mean of 3.56% and multiparous cows had 3.50% (Table 1).
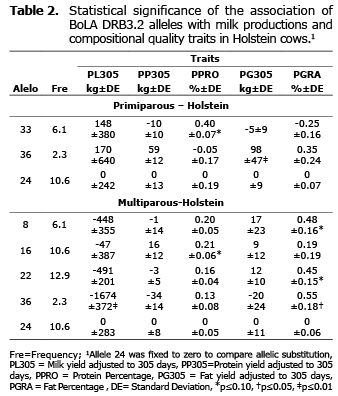
Association of BoLA DRB3.2 alleles with PL305, PP305 and PG305. Comparing the PL305 means of each BoLA DRB3.2 allele with the substitution allele (allele 24), a highly significant association in multiparous cows was found, with the decrease in PL305 from allele 36 with 1674 less than those cows carrying the substitution allele (p≤0.01) (Table 2). A significant association was also found in Holstein cows with a rise in the PG305 for allele 36 (p≤0.01) in primiparous cows. No significant associations were determined for these characteristics in BxH cows.
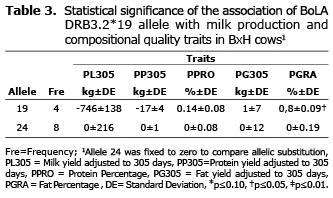
Association of BoLA DRB3.2 alleles with PPRO and PGRA. Significant associations were determined in the Holstein breed with a rise in PPRO for allele 33 in primiparous cows (p≤0.10) with 0.40% more than in cows bearing the substitution allele and for allele 16 in multiparous cows with 0.21% more than in cows bearing the substitution allele (p≤0.10) (Table 2). For PGRA, allele 36 was associated with a rise in the levels of this trait in multiparous cows (p≤0.05), with 0.55% more than in cows that bear the substitution allele. Alleles 8 and 22 were likewise associated to a rise in PGRA in multiparous cows (p≤0.10) (Table 2). For the BxH genetic group, just one significant association was found in allele 19, with a rise in PGRA with 0.8% more than cows that bear allele 24 (p≤0.05), (Table 3).
DISCUSSION
Allelic frequencies of gene BoLA DRB3.2. 27 BoLA DRB3.2 alleles were identified at Paysandú cattle farm in genetic groups Holstein and BxH, indicating a high polymorphism for this gene. The Holstein genetic group had 23 BoLA DRB3.2 alleles, and the 10 most frequent alleles were: BoLA DRB3.2*23, 22, 24, 16, 33, 8, 39, 37, 27 and 18 presenting an accumulated frequency of 0.78. These results were similar to those obtained by various authors who determined the allele frequencies of the gene BoLA DRB3.2 in various Holstein populations and which Support the results of this investigation (6,10,11,17-19).
On the other hand, 18 BoLA DRB3.2 alleles were identified for the BxH genetic group, out of which 11 were reported to be from the blanco Orejinegro (BON) breed, (20). The allelic frequencies obtained in BxH cows in this study were different, compared to those obtained from the BON breed (20). The differences in the allelic frequencies is possibly due to the fact that mainly F2 (25% BON and 75% Holstein) animals were used in this investigation.
Association of BoLA DRB3.2 alleles with production traits. Several BoLA DRB3.2 alleles were reported associated to production traits; mainly milk, fat and protein production (6,10,12,21-23), (Table 4). Significant association found in this study were among alleles 8, 16 and 22, which were also reported by other authors (Table 4). However, some associations from alleles 19, 33 and 36 with production trait determined in this study, have not been reported by other authors.
In this study, allele 8 was significantly associated to an increase in PGRA in multiparous cows of the Holstein genetic group. This result agrees with that reported by Sharif et al (10), who determined significant associations with an increase in fat production. However, those authors, also determined that allele 8 was associated to an increase in milk and protein production in Holstein cows, in contradiction to Starkenburg et al (6), who determined significant associations of allele 8 with a decrease in milk, fat and protein production in cows from the selection line of third lactation. Additionally, allele 8 was associated significantly to a decrease in protein and milk production during the third lactation period, in combined lines (control and selection). Rupp et al (11), likewise, determined significant associations of allele 8 with a decrease in milk production and an increase in protein production (Table 4).
Additionally, during this investigation, a significant association of allele 16 with increase in PPRO in multiparous Holstein cows, Rupp et al (11), determined a significant association of allele 16 with a decrease in the fat production in Holstein cow for the three first lactation periods and Machado et al (21), found for the same allele, a significant association with milk production in Gyr cows. This study found that allele 22 has a significant association with an increase in PGRA in multiparous cows of the Holstein genetic group, which agrees with the results obtained by Pashmi et al (22), who found an increase in fat percentage in Iranian Holstein. Sharif et al (10) did not find associations of allele 22 with fat production or percentage, but did determine significant associations of this allele with a decrease in milk and protein production in both previous and subsequent lactating groups (Table 4).
Allele 36 was another variant of gene BoLA DRB3.2 that was significantly associated with an increase in PG305 in primiparous cows (p<0.01), with an increase in PGRA (p<0.05) and with a decrease in PL305 in multiparous Holstein cows (p<0.01). Another allele that presented significant associations in BxH cows, was allele 19 (absent in Holstein), in this case, it was found to be significantly associated to an increase in PGRA. Associations to productive characteristics or infectious diseases have not yet been reported for these two alleles.
In results obtained by other authors, and compared with those found in this investigation, only one discrepancy was found for allele 8, which was associated with an increase in fat percentage during this study, and which Starkenburg et al (6), determined that was associated with a decrease on fat production. Likewise, there were contradictions between the results obtained by other authors, such as Sharif et al (10) who found that allele 8 was associated to an increase in milk, fat and protein production, while Starkenburg et al (6) found associations of allele 8 with a decrease in milk, fat and protein production. Rupp et al (11), reported associations of allele 8 with a decrease in the production of milk and an increase in the production of protein.
Sharif et al (10), establishes that these inconsistencies may be due to the different states of linkage disequilibrium between populations and that it is also necessary to apply higher statistical astringency to exclude the possibility of presence of erroneous associations, caused by multiple comparison samples. Other factors that may be involved in the inconsistencies among results obtained by different authors, may possibly be the Genotype-Environment interaction, given that the investigations are undertaken in different parts of the world. However, to obtain more precise results, the use of populations with high number of individuals is required, given that the elevated number of polymorphism of gene DRB3, leads to the gaining of low allelic and genotype frequencies.
Rupp et al (11), considers that these differences have been notoriously difficult to avoid and are one of the strong impediment for a selection based on the breeding value in the CMH genotype. However, he explains, by quoting Alizadeh et al (23) that this cannot conclude that the information in the expression of CMH genes, does not improve the mechanisms involved in the resistance to diseases or that it is not one of the many tools that may be used to improve animal health and indirectly productivity in these particular populations.
Gene BoLA DRB3 has been associated to resistance and susceptibility to infectious diseases and its main function is the expression of antigen-presenting proteins (7). However, it is also associated to productive traits. Although the relationship between the gene BoLA with these characteristics is not direct, given that CMH genes are not involved in the process of milk secretion. The relationship is given when finding animal with allele variants that allow them to be resistant to infectious diseases, and this resistance must be reflected by an increase in productivity, or that very productive animals present allelic variants with susceptibility to infectious diseases (11,24).
This investigation found that allele 8 was associated to an increase in the fat percentage in multiparous Holstein cows (p≤0.1). However, other investigations report that it was associated to susceptibility to subclinical mastitis (6,22,25). Likewise, this investigation determined that allele 33 was significantly associated to the decrease in protein percentage in Holstein primiparous cows (p≤0.1), and it was also reported that allele 33 is associated to resistance of subclinical mastitis (25). Even though some alleles have been associated to production and mastitis, it is clear that alleles from gene BoLA DRB3.2 have not yet been reported, to present concluding superiority for productive traits and resistance to infectious diseases. However, the results obtained in this investigation and in those of other authors, generate great interest in finding genes associated to sanity and production, or even other economically important traits, to be included in molecular marker assisted selection programs.
Acknowledgements
This article was part of the project: Correlation between clinical and subclinical mastitis with the genotypic variants of gene BoLA DRB3.2, present in cows and calves of first calving at the milk farm of hacienda Paysandú at the Universidad Nacional de Colombia, QUIPU code 20101006713, financed by the Dirección de Investigación de la Universidad Nacional de Colombia sede Medellín.
REFERENCES
1. Espinal CF, Martínez HJ, González FA. La cadena de lácteos en Colombia: una mirada global de su estructura y dinámica 1991-2005. [En Línea]. 2005 (fecha de acceso 20 abril de 2011); (74). URL disponible en: http://www.agronet.gov.co/www/docs_agronet/2005112162250_caracterizacion_lacteos.pdf.
2. Fedegan. Estadísticas: Producción Colombia. [En Linea]. 2013. (Fecha de acceso 8 de diciembre de 2013). URL disponible en: http://www.fedegan.org.co/estadisticas/produccion-0. [ Links ]
3. Oltenacu PA, Broom DM. The Impact of genetic selection for increased milk yield on the welfare of dairy cows. Animal Welf 2010; 19(S):39-49. [ Links ]
4. Carlén E, Strandberg E, Roth A. Genetic parameters for clinical mastitis, somatic cell score, and production in the first three lactations of Swedish Holstein cows. J Dairy Sci 2004; 87:3062-3070. [ Links ]
5. Quintero GE. Evolución y desarrollo del sector Lácteo en Colombia desde la perspectiva del eslabón primario (Producción). Medellín Colombia: Corporación Universitaria La Sallista, Facultad de Ciencias Administrativas y Agropecuarias. Posgrado en Especialización en Gerencia Agropecuaria: 2011.
6. Starkenburg RJ, Hansen LB, Kehrli JR, Chester-Jones H. Frequencies and effects of alternative DRB3.2 alleles of bovine lymphocyte antigen for Holstein in milk selection and control lines. J Dairy Sci 1997; 80:3411-3419. [ Links ]
7. Behl JD, Verma NK, Tyagi N, Mishra P, Behl R, Joshi BK. The major histocompatibility complex in bovines: A Review. ISRN Veterinary Science 2012; 2012:1-12. [ Links ]
8. Wang K, Sun D, Zhang Y. Sequencing of 15 new BoLA-DRB3 Alleles. Int J Immunogenet 2008; 35:331-332. [ Links ]
9. Lee BY, Hur TY, Jung YH, Kem H. Short Communication. Identification of BoLA DRB3.2 alleles in Korean native cattle (Hanwoo) and Holstein populations using a next Generation sequencer. Anim Genet 2012; 43(4):438-441. [ Links ]
10. Sharif S, Mallard BA, Wilkie BN, Sargeant JM, Scott HM, Dekkers JC et al. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with production traits in Canadian dairy cattle. Anim Genet 1998; 30:157-160. [ Links ]
11. Rupp R, Hernandez A, Mallard BA. Association of bovine leukocyte antigen (BoLA) DRB3.2 with immune response, mastitis and production and type traits in Canadian Holsteins. J Dairy Sci 2007; 90:1029-1038. [ Links ]
12. Nascimento CS, Machado MA, Martínez ML, Barbosa MV, Martins MF, Campos AL, et al. Association of the bovine major histocompatibility complex (BoLA) BoLA-DRB3 gen with fat and protein production and somatic cell score in Brazilian Gyr Dairy cattle (Bos indicus). Genet Mol Biol 2006; 29(4):641-647. [ Links ]
13. Wojdak-Maksymiec K, Kmiec M, Kowalewska-Luczac I, Warliñski M. DRB3 Gene polymorphism and somatic cell count in milk of jersey. J Anim Vet Adv 2010; 9(9):1295-1300. [ Links ]
14. Van Eijk MJT, Stewart-Haynes JA, Lewin HA. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim Genet 1992; 23:483. [ Links ]
15. Laird NM, Lange C. The Fundamentals of Modern Statistical Genétics. New York USA: Springer; 2011.
16. SAS/STAT: [Programa de ordenador] Version 9.1, Cary (NC): SAS Institue Incorporation; 2006.
17. Nassiry MR, Shahroodi EF, Mosafer J, Mohammadi A, Manshad E et al. Analysis and frequency of bovine lymphocyte antigen (BoLA DRB3) alleles in Iranian Holstein Cattle. Russ J Genet 2005; 41(6):664-668. [ Links ]
18. Nassiry MR, Sadeghi B, Tohidi R, Afshari JT, Khosravi M. Comparison of bovine lymphocyte antigen DRB3.2 allele frequencies between two subpopulations of Iranian Holstein cattle. Afr J Biotechnol 2008; 7(15):2671-2675. [ Links ]
19. Operzadek J, Urtnowski P, Sender G, Pawlik A, Lukaszewicz. Frequency of alleles BoLA-DRB3 in Polish Holstein-Friesian cattle. Anim Sci Pap Rep 2012; 30(2):91-101. [ Links ]
20. Martínez R, Toro R, Montoya F, Burbano M, Tobón J, Gallego J y Ariza F. Caracterización del locus BoLA-DRB3 en ganado criollo colombiano y asociación con resistencia a enfermedades. Arch Zoot 2005; 54:(206-207):350-356. [ Links ]
21. Machado MA, Nascimento CS, Martinez ML, Silva GB, Campos AL, Teodoro RL, et al. Association of BoLA-DRB3.2 locus with bovine milk production in Gyr breed. Arq Bras Med Vet Zootec 2005; 57(3):380-389. [ Links ]
22. Pashmi M, Qanbari S, Ghorashi SA, Sharifi AR. Analysis of relationship between bovine lymphocyte antigen DRB3.2 alleles, somatic cell count and milk traits in Iranian Holstein population. J Anim Breed Genet 2009; 126:296-303. [ Links ]
23. Alizadeh Z, Karrow N, Mallard BA. Biological effect of varying peptide binding affinity to the BoLA-DRB3*2703 allele. Genet Sel Evol 2003; 35(1):51-65. [ Links ]
24. Rupp R, Boichard D. Genetics of resistance to mastitis in dairy cattle. Vet Res 2003; 34:671-688. [ Links ]
25. Zambrano JC, López-Herrera A, Echeverri JJ. Alleles of the BoLA DRB3.2 gene are associated with mastitis in dairy cows. Rev Col Cienc Pec 2011; 24(2):145-156. [ Links ]