Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista MVZ Córdoba
Print version ISSN 0122-0268
Rev.MVZ Cordoba vol.20 supl.1 Córdoba Dec. 2015
ORIGINAL
Pseudomonas aeruginosa: an emerging nosocomial trouble in veterinary
Pseudomonas aeruginosa: un problema nosocomial emergente en veterinaria
Yuly Bernal-Rosas,1 M.Sc, Karen Osorio-Muñoz,1 M.V, Orlando Torres-García,1* Ph.D.
1 Universidad Antonio Nariño, Facultad de Medicina Veterinaria, Carrera 3 este # 47A-15, Bogotá, D.C., Colombia.
*Correspondence: ortorres@uan.edu.co
Received: August 2014; Acepted: February 2015.
ABSTRACT
Objective. The goal of this study was to evaluate the susceptibility pattern of isolates P. aeruginosa from veterinary clinical centers in Bogotá, D.C., to some commonly used antibiotics in clinical. Materials and methods. Bacteriological standard protocols were used for the isolation and identification of bacterial strains. To evaluate the antimicrobial susceptibility of the isolates, to commonly used antibiotics, was performed the Kirby-Bauer agar-disk diffusion method on Mueller-Hinton agar. Results. A total of 160 samples was taken from clinical specimens and the environment in different veterinary clinics. Out of these samples, 89 (55.6%) were gram-negative strains, of which ten strains of P. aeruginosa were isolated (11.2%). All strains were resistant to Cefazolin, Lincomycin, Cephalothin, Ampicillin, Clindamycin, Sulfamethoxazole-Trimethoprim and Chloramphenicol while some isolates exhibited either resistance or an intermediate response to Amikacin (30%), Gentamicin (30%), Tobramycin (10%), Ciprofloxacin (20%), Ceftazidime (30%), Erythromycin (100%), Tetracycline (100%), Imipenem (10%), Meropenem (90%) and Bacitracin (90%). Conclusions. The results demonstrate that the acquired antimicrobial resistances of P. aeruginosa strains depend on antibiotic protocols applied. As observed in human hospitals, Pseudomonas aeruginosa is acting as one of the multidrug-resistant microorganisms of veterinary clinical relevance.
Key words: Antibiotic resistance, Bacteria, Dog, Multidrug-resistant, Nosocomial infections. (Source: NLM).
RESUMEN
Objetivo. Evaluar el comportamiento a algunos antibióticos de uso común en clínica de cepas de Pseudomonas aeruginosa aisladas de centros clínicos veterinarios en Bogotá, D.C. Materiales y métodos. Un protocolo bacteriológico estándar se utilizó para el aislamiento e identificación de cepas bacterianas. Para evaluar la sensibilidad de los aislamientos bacterianos a diferentes antimicrobianos de uso clínico común, se empleó el método de difusión sobre agar Mueller-Hinton de Kirby-Bauer. Resultados. Un total de 160 muestras fueron tomadas a partir de muestras clínicas y del ambiente de diferentes clínicas veterinarias de la ciudad de Bogotá. De estas muestras, 89 (55.6%) fueron cepas bacterianas gram-negativas, de las que se aislaron 10 cepas de P. aeruginosa (11.2%). Todas las cepas de P. aeruginosa mostraron resistencia a la Cefazolina, Lincomicina, Cefalotina, Ampicilina, Clindamicina, Trimetoprim- Sulfametoxazol y Cloranfenicol; mientras que algunas de éstas, mostraron resistencia o una respuesta intermedia a Amikacina (30%), Gentamicina (30%), Tobramicina (10%), Ciprofloxacina (20%), Ceftazidima (30%), Eritromicina (100%), Tetraciclina (100%), Imipinem (10%), Meropenem (90%) y Bacitracina (90%). Conclusiones. Los resultados demuestran que la resistencia adquirida a los antimicrobianos en las cepas de P. aeruginosa aisladas de estos centros clínicos depende de los protocolos antibióticos aplicados. Al igual que sucede en los nosocomios Humanos, Pseudomonas aeruginasa se está comportando como uno de los microorganismos multirresistetes de relevancia clínica causante de infecciones nosocomiales veterinarias.
Palabras clave: Bacterias, infecciones nosocomiales, multiresistencia, perros, resistencia a antibioticos. (Fuente: NLM).
INTRODUCTION
Resistance to antibiotics is a growing problem worldwide, especially among nosocomial pathogens, and the spread of resistant bacteria is favored by the inappropriate use of antibiotics (1), leading to longer hospital stays and higher treatment costs (2). Several studies suggest that the antibiotic resistance is increasing from nosocomial isolates (3). The nutritional versatility and the ubiquity of Pseudomonas aeruginosa have made it becomes one of the most-common causes of Gram-negative nosocomial infections (4).
Pseudomonas aeruginosa is a motile, gram-negative rod that belongs to the family Pseudomonadaceae. P. aeruginosa is widely distributed in nature, and it has been isolated from various biological samples and different inorganic surfaces (5,6). P. aeruginosa is increasingly recognized as an emerging opportunistic pathogen of clinical relevance that causes of chronic and recurrent infections in both humans and animals, particularly, in dogs and cats the most-frequent diseases include otitis external/media, urinary tract infection and pyoderma (7-9).
In Latin America, bacterial resistance in hospitals presents a high variety with small differences among countries, including Colombia. Epidemiological studies conducted in Bogotá in early 2000 showed a prevalence of 37% for P. aeruginosa, this being one of clinically relevant bacteria that most commonly cause nosocomial infections reviewed by Cabrera et al (10). However, no studies have analyzed the impact of this problem in veterinary clinical centers, in Bogotá.
P. aeruginosa isolates are naturally resistant to a large number of antibiotics; however, mechanisms of antimicrobial resistance, such as efflux pumps and porins, has allowed to these strains extend the range of antibiotic resistance (11). Multiple antibiotic resistances have created significant problems in the treatment of infected wounds. On the other hand, treatment failure and cross-resistance between strains and the spread of strains from patient to patient can result in multidrug resistance (MDR) P. aeruginosa. (12).
Taking into account the above considerations, the aim of this survey was to assess the susceptibility pattern of P. aeruginosa isolates from veterinary clinical centers in Bogotá, D.C., to some commonly used antibiotics in clinical.
MATERIALS AND METHODS
Study design. This prospective observational study was carried out at the Veterinary Faculty of Antonio Nariño University from Bogotá, D.C., Colombia, between June 2013 and May 2014. Dog patients that were considered to be at high risk of developing nosocomial events were eligible for enrollment in the study (Most hospitalized patients presented primarily infections of viral or bacterial type). The study was restricted to patients (canines) that were hospitalized for at least one day in the respective critical care units (88.9% of patients were hospitalized for more than 4 days).
Bacterial isolates. 135 samples from a variety of clinical specimens of dogs (stool (n =91), urine (n =14), nasal discharge (n =6), wound swab (n =8), surgical site infections (n =3), swab from skin (n =8) and ear (n =5)) and 25 environmental samples (surgical instruments, dressings, baths, surgical rooms, devices, floors, walls, beds, sinks, examination tables and wastewater) were examined. Cotton swabs were used to collect samples. The samples were placed into Brain Heart Infusion (BHI) broth (Oxoid, UK) and were sent immediately to the microbiology laboratory.
Sample processing. Samples were inoculated onto 5% of sheep blood agar plates (Oxoid, UK) and incubated at 37°C for 24 h. The identification of Pseudomonas was on the basis of colony morphology and Gram Staining; any suspicious colony was subcultured on cetrimide agar plates (CM0579 Oxoid, UK), and purified. The isolate were identified as a P. aeruginosa based on biochemical test. Such as oxidase test, motility, aerobic fermentation of glucose, lactose, maltose and mannitol, arginine and lysine utilization, nitrate reduction, production of urease and DNAse, ONPG (orthonitrophenyl-beta-D-galactopyranoside) test, acetamide and esculin hydrolysis, and susceptibility to polymyxin, was performing according to (13).
Isolates were stored at -80°C in Mueller-Hinton Broth (Merck, Germany) supplemented with 40 per cent glycerol (v/v).
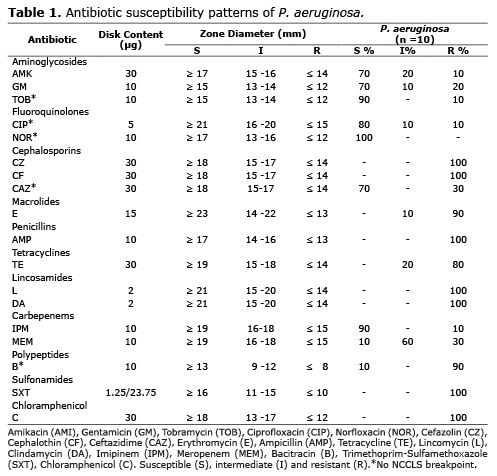
Antibiotic sensitivity assay. Antimicrobial susceptibility to a panel of 18 antimicrobial agents most commonly used in veterinary clinical centers in Bogota was carried out on Muller-Hinton agar by using Kirby Bauer disc diffusion technique reformation by the Performance Standards for Antimicrobial Disk Susceptibility Tests (CLSI) suggestions (14). The following antimicrobials for P. aeruginosa were used: Aminoglycosides: Amikacin (AMI 30 µg), Gentamicin (GM 10 µg) and Tobramycin (TOB 10 µg). Fluoroquinolones: Ciprofloxacin (CIP 5 µg) and Norfloxacin (NOR 10 µg). Cephalosporins: First generation: Cefazolin (CZ 30 µg) and Cephalothin (CF 30 µg); Third generation: Ceftazidime (CAZ 30 µg). Macrolides: Erythromycin (E 15 µg). Penicillins: Ampicillin (AMP 10 µg). Tetracyclines: Tetracycline (TE 30 µg). Lincosamides: Lincomycin (L 2 µg) and Clindamycin (DA 2 µg). Carbepenems: Imipinem (IPM 10 µg) and Meropenem (MEM 10 µg). Polypeptides: Bacitracin (B 10 µg). Sulfonamides: Trimethoprim-Sulfamethoxazole (SXT 1.25/23.75 µg). Chloramphenicol: Chloramphenicol (C 30 µg).
The inhibition zone diameter around each disk was measured and was interpreted as described by the NCCLS, which classifies the organism into three categories: sensitive (S), intermediate sensitive (I) and resistant (R) (14).
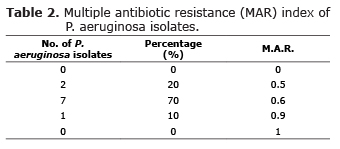
Multiple antibiotic resistance index. The multiple antibiotic resistance (MAR) index was determined for each isolate by dividing the number of antibiotics to which the isolate is resistant by the total number of antibiotics tested (15). Multidrug resistance was defined as resistance to two or more antimicrobial classes. Any sign of inhibition or sensitivity was considered to be indicative of nonresistance.
Quality Control of antimicrobial susceptibility tests. Quality control was performed utilizing strains from the American type culture collection (ATCC), P. aeruginosa ATCC 27853.
Ethical approval. Regarding ethical considerations, these studies adhered to the Article 11 from Resolution No. 008430 on October 4, 1993 by the Colombian Ministry of Health according to whose classification, these are minimum-risk studies.
RESULTS
Between February 2013 and June 2013, 160 bacterial strains were collected from a variety of clinical specimens and environment samples of several veterinary clinics. Out of these samples, 89 (55.6%) were gram-negative strains, of which 10 (11.2%) strains of P. aeruginosa were isolated. 3 (30%) were isolated from environmental samples, and 7 (70%) were isolated from clinical specimens.
In this study, P. aeruginosa was most commonly isolated from surgical sites (57.1%). The percentage of strains susceptible, intermediate, or resistant to each antibiotic is presented in table 1. All 10 (100%) strains tested were susceptible to Norfloxacin, followed by Tobramycin (90%), Imipinem (90%), Ciprofloxacin (80%), Amikacin (70%), Gentamicin (70%), Ceftazidime (70%). Of the remaining antibiotics tested, Meropenem and Bacitracin exhibited the least activity against P. aeruginosa with 90% of the resistant strains each one. Notably, 100% of the isolates exhibited resistance to Cefazolin, Lincomycin, Cephalothin, Ampicillin, Clindamycin, Sulfamethoxazole-Trimethoprim and Chloramphenicol while some isolates exhibited either resistance or an intermediate response to Amikacin (30%), Gentamicin (30%), Tobramycin (10%), Ciprofloxacin (20%), Ceftazidime (30%), Erythromycin (100%), Tetracycline (100%), Imipinem (10%), Meropenem (90%) and Bacitracin (90%).
Multiple antibiotic resistances were seen in all Pseudomonas aeruginosa isolated strains. However, one of these clinic samples, showed the highest MDR 16/18 antibiotics tested (MAR 0.9). Table 2 shows multiple antibiotic resistance (MAR) index of P. aeruginosa isolates, which shows that, 80% had MAR index of 0.5 and above.
DISCUSSION
Pseudomonas aeruginosa is a major cause of nosocomial infections in immunocompromised patients, especially in hospitalized burn patients (16). P. aeruginosa is intrinsically resistant to several antibiotics because of the low permeability of its outer-membrane, the constitutive expression of various efflux pumps, and the production of antibiotic-inactivating enzymes (11).
Infections caused by resistant strains are a true concern in many hospitals worldwide since they are associated with a high mortality, secondary bacteremia, an increase in the length of hospital stay, and a considerable increase in healthcare costs (17).
Has been reported that about 5 to 10% of hospitalized human patients acquire an infection of hospital origin (18), not very dissimilar from the observed percentages in veterinary practice in small animal species, where prevalence reported is between 0.8 and 18.1%, according to the established surgical procedure (19). So the present study was conducted to determine the antibiotic resistance pattern of Pseudomonas aeruginosa isolated from clinical and environmental samples in various veterinary clinical centers in Bogotá, D.C.
In the present study, the highest rate (51.7%) of Pseudomonas aeruginosa infections was observed in the examination room and surgical ward, followed by isolating strains of environment, in clinical floors, surgical rooms and exam table (30%). These results are in line with studies of Rakesh on 2012 (20).
Aminoglycosides kill bacteria by inhibiting protein synthesis via binding to the 16S rRNA and by disrupting the bacterial cell membrane integrity (21). In this study, ten percent P. aeruginosa is intermediate or marginally sensitive to gentamicin with 20% resistant these data reveal that the gentamicin resistance is increasing. These results are similar to those reported in previous studies (15). However, the newer aminoglycosides with a broader spectrum of antibacterial activity, such as Amikacin, Gentamicin and Tobramycin, which in this study, showed relatively high activity of 70%, 70% and 90% against P. aeruginosa strains, respectively. Similarly, Harada and Lin, reported on 2012, a low percentage of resistance to Amikacin and Gentamicin (22, 23). Suggesting that these could still be the gold standard for treatment in cases of more resistant bacteria.
Fluoroquinolones, particularly Ciprofloxacin, are commonly used in the treatment of P. aeruginosa infections. Ciprofloxacin is the one antibiotic with antipseudomonal coverage. P. aeruginosa is most susceptible to Ciprofloxacin and Imipenem (16, 24). In contrast, this study shows that these isolates have started developing resistance or marginally sensitive (20%) against Ciprofloxacin, probably due to its irrational and inappropriate use. These results are consistent with those reported in other studies (22, 25, 26).
Was found that strains of P. aeruginosa isolates are resistant to multiple drugs, suggesting that selective pressure created by the persistent and possibly inappropriate use of these antimicrobials in veterinary clinic practice is one of the determining factors for the emergence of resistance; reason that justifies the MIR greater than 0.5 in 80% of P. aeruginosa strains isolated in the study; these results are consistent with those reported by Kyung-Mee on 2013 (27).
β-Lactam antibiotics, including antipseudomonal Penicillins, Cephalosporins, Monobactams, and Carbapenems, remain essential components for the treatment of nosocomial infections by Pseudomonas aeruginosa (28). Nevertheless, in this study, with the exception of Ceftazidime and Imipinem, all P. aeruginosa strains, were resistant to Cefazolin, Cephalothin, Ampicillin and Meropenem. In the same way, other antibiotics probed in this study such as, Macrolides, Tetracyclines, Lincosamides, Polypeptides, Sulfonamides and Chloramphenicol, exhibited the least activity against P. aeruginosa; resistance to these first-line antibiotics is increasing and frequently associated with multidrug resistance (MDR) phenotypes (29, 30). These results could suggest the possible extended-spectrum beta-lactamase (ESBL) production in these MDR strains of P. aeruginosa; nonetheless, confirmation studies are required.
Numerous reasons can be assigned to the development of antibiotic resistant strains. In developing countries due to self-medication, free sale of antibiotics and antibiotic prescribing broad-spectrum that are activity against a wide range of different microbes, are the main sources of induction of antimicrobial resistance.
In conclusion the nosocomial infections in veterinary medicine are an emerging trouble.
Multidrug resistance was observed in all strains of P. aeruginosa isolated in this work, possibly is due to antibiotic protocols applied in veterinary clinical. Like to in human hospitals, Pseudomonas aeruginosa today, is one of the microorganisms of veterinary clinical relevance in Bogota.
These results highlight the need to implement microbiological monitoring programs for multidrug resistant organisms and infection control procedures at veterinary clinical centers. Additionally, the mandatory incorporation of antibiograms as a guide in selecting empiric antibiotic therapy. Finally, the possibility of bacterial spread from animals to humans or vice versa requires particular attention.
Conflict of interest statement
The authors do not have any financial or other associations that might pose a conflict of interest.
Acknowledgements
This work was financially supported by a grant from the University Antonio Nariño.
REFERENCES
1. W.H.O. The evolving threat of antimicrobial resistance - Options for action. Geneva. W.H.O.; 2012. [ Links ]
2. Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis 2007; 13(12):1840-1846. [ Links ]
3. Dai CM, Zhou JD, Wu Y, Qi Y. Distribution of pathogen and resistance of nosocomial infections in the intensive care units. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2006; 31(2):277-280. [ Links ]
4. Obritsch MD, Fish DN, MacLaren R, Jung R. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy 2005; 25(10):1353-1364. [ Links ]
5. Park HR, Hong MK, Hwang SY, Park YK, Kwon KH, Yoon JW, et al. Characterisation of Pseudomonas aeruginosa related to bovine mastitis. Acta Vet Hung 2014; 62(1):1-12. [ Links ]
6. Morales E, Cots F, Sala M, Comas M, Belvis F, Riu M, et al. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Serv Res 2012; 12:122. [ Links ]
7. Hariharan H, Coles M, Poole D, Lund L, Page R. Update on antimicrobial susceptibilities of bacterial isolates from canine and feline otitis externa. Can Vet J 2006; 47(3):253-255. [ Links ]
8. Hall JL, Holmes MA, Baines SJ. Prevalence and antimicrobial resistance of canine urinary tract pathogens. Vet Rec 2013; 173(22):549. [ Links ]
9. Papich MG. Antibiotic treatment of resistant infections in small animals. Vet Clin North Am Small Anim Pract 2013; 43(5):1091-1107. [ Links ]
10. Cabrera CE, Gómez RF, Zuñiga AE, Corral RH, Bertha López B, Chávez M. Epidemiology of nosocomial bacteria resistant to antimicrobials. Colombia Médica 2011; 42(1):117-125. [ Links ]
11. Hillier A, Alcorn JR, Cole LK, Kowalski JJ. Pyoderma caused by Pseudomonas aeruginosa infection in dogs: 20 cases. Vet Dermatol 2006; 17(6):432-439. [ Links ]
12. Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother 2006; 50(1):43-48. [ Links ]
13. Washington C. Winn SDA, William M. Janda, Elmer W. Koneman, Paul C. Schreckenberger, Gary W. Procop and Gail L. Woods. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2006. [ Links ]
14. C.L.S.I. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard - Fourth Edition. CLSI document, VET01-A4. Wayne, PA: Clinical and Laboratory Standards Institute; 2013. [ Links ]
15. Tripathi P, Banerjee, G., Saxena, S., Kumar, G.M., and Ramteke, P.W. Antibiotic resistance pattern of Pseudomonas aeruginosa isolated from patients of lower respiratory tract infection. Afr J Microbiol Res 2011; 5(19):2955-2959. [ Links ]
16. Biswal I, Arora BS, Kasana D, Neetushree. Incidence of multidrug resistant Pseudomonas aeruginosa isolated from burn patients and environment of teaching institution. J Clin Diagn Res 2014; 8(5):DC26-29. [ Links ]
17. Giamarellou H. Prescribing guidelines for severe Pseudomonas infections. J Antimicrob Chemother 2002; 49(2):229-233. [ Links ]
18. Sydnor ER, Perl TM. Hospital epidemiology and infection control in acute-care settings. Clin Microbiol Rev 2011; 24(1):141-173. [ Links ]
19. Nelson LL. Surgical site infections in small animal surgery. Vet Clin North Am Small Anim Pract 2011; 41(5):1041-1056. [ Links ]
20. Rakesh RM GN, Kalpesh M, Rosy P, Kanu P, Vegad MM. Antibiotic resistance pattern in Pseudomonas aeruginosa species isolated at a tertiary care hospital, ahmadabad. Natl J Med Res 2012; 2(2):156 - 159. [ Links ]
21. Shakil S, Khan R, Zarrilli R, Khan AU. Aminoglycosides versus bacteria--a description of the action, resistance mechanism, and nosocomial battleground. J Biomed Sci 2008; 15(1):5-14. [ Links ]
22. Harada K, Arima S, Niina A, Kataoka Y, Takahashi T. Characterization of Pseudomonas aeruginosa isolates from dogs and cats in Japan: current status of antimicrobial resistance and prevailing resistance mechanisms. Microbiol Immunol 2012; 56(2):123-127. [ Links ]
23. Lin D, Foley SL, Qi Y, Han J, Ji C, Li R, et al. Characterization of antimicrobial resistance of Pseudomonas aeruginosa isolated from canine infections. J Appl Microbiol 2012; 113(1):16-23. [ Links ]
24. Hirakawa Y, Sasaki H, Kawamoto E, Ishikawa H, Matsumoto T, Aoyama N, et al. Prevalence and analysis of Pseudomonas aeruginosa in chinchillas. BMC Vet Res 2010;6:52. [ Links ]
25. Hu XH, Xu XM, Mi ZH, Fan YF, Feng WY. Relationship between drug resistance of Pseudomonas aeruginosa isolated from burn wounds and its mobile genetic elements. Zhonghua Shao Shang Za Zhi 2009; 25(2):103-105. [ Links ]
26. Ali SQ, Zehra A, Naqvi BS, Shah S, Bushra R. Resistance pattern of ciprofloxacin against different pathogens. Oman Med J 2010; 25(4):294-298. [ Links ]
27. Park KM, Nam HS, Woo HM. Successful management of multidrug-resistant Pseudomonas aeruginosa pneumonia after kidney transplantation in a dog. J Vet Med Sci 2013; 75(11):1529-1533. [ Links ]
28. Mesaros N, Nordmann P, Plesiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, et al. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 2007; 13(6):560-578. [ Links ]
29. Kiser TH, Obritsch MD, Jung R, MacLaren R, Fish DN. Efflux pump contribution to multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Pharmacotherapy 2010; 30(7):632-638. [ Links ]
30. Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodriguez C, Moya B, et al. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 2011; 55(5):1906-1911. [ Links ]