INTRODUCTION
Infectious bronchitis virus (IBV) is the causative agent of infectious bronchitis (IB), an acute and highly contagious in poultry, which primarily affects respiratory tract, but different IBV strains may show variable tissue tropisms and also affect the reproductive, respiratory and renal tract (1). Infected poultry become predisposed to secondary infections, and in this case it causes an increase in mortality rate in breeding 2. The disease has worldwide distribution, being one of the biggest problems of the poultry industry, affecting the performance of laying hens and broilers and expenses to control the disease 2,3.
Both inactivated and live attenuated vaccines have been used to control the IB, nonetheless some outbreaks continue to happen 3. This control is not being effective because of the lack of cross-protection to the large number of variants and serotypes that are emerging. Studies using molecular methods have shown that the new strains or serotypes have only a few changes in amino acid in the S1 portion of the viral spike protein, while most of the genome remains unchanged 4,5. These changes may occur due to an immune pressure caused by widespread use of vaccines, genetic recombination or of coexistent infections, or even a dominant serotype reduction as a result of vaccination, allowing other emerging field strains 1,4,6,7. The transmembrane domain is one of cleavage S1 region of the S virus structural protein, which is a hypervariable region, and the most antigenic and responsible for inducing protection against the virus 3.
In Colombia, vaccines with Massachusetts (Mass), Connecticut (Conn) e Arkansas strains are permitted to marketing 6, and even then, the disease is not being controlled, probably because the vaccine strain does not induce cross-protection to different genotypes that have emerged in the field with point mutations and deletion events insertion in the hypervariable region S1 4. Other countries such as Brazil, Argentina, Australia and Tunisia also have the same situation. Vaccine strategies using licensed vaccines to control the IB have been ineffective, due to the circulation of new genotypes 7-12. Therefore, after identifying the IBV field genotypes it is important to implement appropriate control plans, using specific vaccine strains for each region in order to offer protection against the disease.
The aim of this study was to evaluate the different genotypes of IBV in commercial poultry flocks from different farms of the Tolima Department, in Colombia and their relationship with reference strains, including some vaccine strains allowed in the country.
MATERIALS AND METHODS
Samples. Twenty-one farms of seven different municipalities (Mariquita, Piedras, Ibagué, Coello, Valle de San Juan, San Luis and Guamo) of the Tolima Department have been selected to carry out the sample collection. Samples were taken the rules and ethics committee approval of the Universidad del Tolima. The farms were chosen where poultry had some characteristic respiratory signs. All poultry were vaccinated with live attenuated vaccine containing the Massachusetts strain (M41) (Table 1). Five samples of tracheal swabs of poultry were taken, at random, in each farm. The swabs were placed in special tubes containing 3 ml universal transport medium used for the preservation and transportation of virus (UTM-RT) (Puritan Medical Products, Guilford). After collection the samples, they were frozen at -30 °C. Pools of five samples were prepared to each farm, twenty-one pools in total. The samples were centrifuged at 1800 rpm at 4°C for 10 minutes and the supernatant was collected for analysis. The supernatant was used for identification of positive samples and later for virus isolation.
Reference strains. The GeneBank accession numbers of IBV strains used in this study are: Massachusetts (M41) (AY561711.1); Massachusetts Holland 120 (H120) (GU393335); Massachusetts Holland 52 (H52) (EU817497); Connecticut (Conn 46) (FJ904717); Arkansas DPI (GQ504720); and China QXIBV (AF193423).
The isolated from Brazil 13, Cuba 14 and Argentina 10 were also used to evaluate the phylogenetic relationship with the samples from Colombia. The GeneBank accession numbers of IBV variants are: IBV/Brazil/SC02 (GQ169247), IBV/Brazil/PR07 (GQ169244), IBV/Brazil/SP02 (GQ169250), Cuba/La Habana/CB6/2009 (HE590762), Cuba/La Habana/CB13/2009 (HE590763), Cuba/La Habana/CB19/2009 (HE590764), AR03BA06 (FJ167386), AR06BA13 (FJ167376), AR06BA14 (FJ167375).
RNA extraction. Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions 15, directly of the supernatant of the samples suspensions and of the allantoic fluid of embryonated eggs to negative control (ultrapure water).
cDNA synthesis. After extraction, RNA was submitted to cDNA synthesis by using the reverse transcriptase technique (RT). To each reaction 0.5 μL of RNA was added, 0.25 μL of Super Script II enzyme (Invitrogen - 200U/ μL), 0.75 μL of each primer (20 pmol/μL), 0.13 μL of inhibitor RNAse (40U/μL), 0,5 μL of DL-Dithiothreitol (DTT Thermo Scientific- 0,1 M), 0.25 μL of dNTPs (10mM), 1 μL of Buffer (5x) and 2.12 μL of ultrapure water to complete the final volume of 5.5 μL reaction. The samples were taken to thermocycler following the thermal profile one cycle at 42°C /60 minutes for enzyme activation, one cycle of 70°C /15 minutes for inactivate the reaction and one cycle of 4°C /∞.
Virus screening. To make the identification of positive samples was performed Nested reverse transcriptase-polymerase chain reaction (Nested RT-PCR) to amplifying the partial region of the S1 subunit. The initial PCR used primers SX1+ [5'-CAC CTA GAG GTT TGT TAG CAT G -3'] and SX2- [5'-TCC ACC TCT ATA AAC ACC TTT AC -3']. The amplicon was further amplified in a second internal PCR that used primers SX3+ [5'-TAA TAC TGG CAA TTT TTC AGA TGG -3'] and SX4- [5'-AAT ACA GAT TGC TTA CAA CCA CC -3'] 16.
Amplification was performed in two consecutive reactions. In the first, the RT-PCR was carried out in a total volume of 25 μL, using 5 μL of cDNA, 11.5 μL of highly pure water, 5 μL of Buffer Green (5x), 1.5 μL of MgCl2 (25mM), 0.5 μL of each primer (20 pmol/ μL) SX1+ and SX2-, 0.5 μL de dNTPs (10 mM), 0.25 μL of Taq DNA polymerase (5 U/mL). Amplification was performed in a Thermocycler with the following conditions: one cycle at 94ºC for 15 sec and 35 cycles at 94ºC for 10 sec, 50ºC for 20 sec, and 72ºC for 40 sec, and one cycle at 4ºC/ ∞.
The second PCR (Nested) was carried out in a volume of 25 μL using 1 μL of amplified product, 15.5 μL of highly pure water, 5 μL of Buffer Green (5x), 1.75 μL of MgCl2 (25 mM), 0.5 of each primer (20 pmol/ μL) SX3+ and SX4-, 0.5 μL de dNTPs (10 mM), 0.25 μL of Taq DNA polymerase (5 U/mL). A Thermal cycle was performed with the same equipment following these steps: one cycle at 94ºC for 15 sec and 35 cycles at 94ºC for 10 sec, 50ºC for 20 sec, and 72ºC for 40 sec, and one cycle at 4ºC/ ∞. Amplification was verified by electrophoresis 1.5% agarose gel in 1 x TBE buffer (2 mM of EDTA, 90 mM of Tris-Borate, pH 8.3), using a 100bp ladder as a molecular weight marker for confirmation of the length of the PCR products. Gels were stained with ethidium bromide (0.2 μg/mL).
Virus isolation. The technique was performed according to Owen et al 17 protocol for IBV isolation in embryonated eggs SPF (Specific Pathogen Free). Briefly, the supernatants of IBV-positive samples (as determined by virus screening) were filtered through 0.45 μm; 0.1 ml was inoculated into four 9-day-old specific-pathogen-free embryonated chicken eggs (ICA, Bogotá, Colômbia) by the allantoic cavity route. Eggs were incubated at 37ºC and candled on a daily basis to check for embryo viability. Allantoic fluids were harvested 72 h post inoculation and two further blind serial passages were performed in a similar way. Each passage (allantoic fluid) was tested for the presence of IBV by Nested RT-PCR described above.
DNA sequencing and phylogenetic analysis. Sequencing was performed in Corporación CorpoGen (CorpoGen Investigación y Biotecnología, Bogotá, Colombia). It was used 20 μL of amplicon unpurified corresponding to the S1 region. In the Corporation CorpoGen, the product was submitted to purification by alcoholic precipitation 18 for removal of small molecules as dNTPs and primers not annealed. After the purification process, samples were analyzed on an agarose gel to verify the presence of a single band without drags. For sequencing it was used the BigDye Terminator v3.1 Cycle Sequencing kit using capillary electrophoresis and the sequencer ABI3730XL according to the manufacturer's instructions. The nucleotides (nt) sequences were edited and aligned with the program Clustal W 28, through Bioedit and analyzed Phylogenetic Analysis Using Parsimony and Other Methods (PAUP) 19 with 2000 bootstrap replicates. The sequences S1 of the isolates and of the six reference sequences retrieved from GeneBank were aligned through the Neighbor-Joining method 20. The evolutionary distances were computed using the p-distance method 21 and they are in the units of the number of base differences per site.
RESULTS
Of 21 samples, four were positive for IBV (HT6, HT9, HT10 and HT11), three positive samples isolated in Ibagué and one in Coello. The field IBV isolates obtained from tracheal swabs of the broilers with respiratory disease symptoms are presented in table 1.
Table 1 Field IBV isolates obtained from tracheal swabs of the broilers with respiratory disease symptoms
| Farm | Isolate | Municipality | Sector | GenBank | Age vaccination* |
|---|---|---|---|---|---|
| A | HT6 | Ibagué | Chucuní | En6Chu14mVa | 1st day |
| B | HT9 | Ibagué | Ambalá | En9Am10mVa | 10th day |
| C | HT10 | Ibagué | Chapetón | En10Cha15mVa | 1st day |
| D | HT11 | Coello | Cocora | En11Coe14mVa | 1st day |
| *Age of the poultry vaccinated with attenuated live vaccine containing serotype M41 Massachusetts |
Virus isolation in chicken embryos for positive samples produced lesions characteristic of IBV, such as curled, hemorrhagic and stunted embryos.
The allantoic fluid of embryonated SPF eggs infected with IBV samples were harvested and submitted to RNA extraction and thereafter cDNA synthesis. Nested RT-PCR was performed to amplify the product of S1 region with 392 bp. The amplified region was confirmed by agarose gel and the product of the nested RT-PCR was used for sequencing of the samples.
The partial region of the S1 subunit of the four isolates was compared with the sequences of reference strains Massachusetts M41, H52 and H120, 46 Connecticut, Arkansas DPI and China QXIBV (Figure 1). The similarity of the isolates and reference strains is shown in table 2.
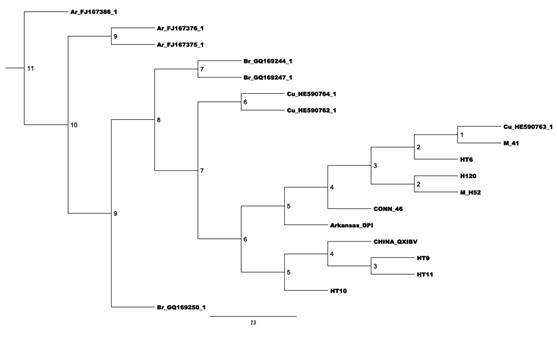
Figure 1 Phylogenetic relationships of the Colombia IBM samples(HT6, HT9, HT10, HT11) and some reference strains bases on S1 partial nt Numbers along the branches refer to bootstrap values.
Table 2. Similarity analysis of nucleotide from the S1 gene of Colombian isolates and reference strains
| Percent nucleotide similarity | ||||||||||
| HT6 | HT9 | HT10 | HT11 | M41 | H120 | Conn46 | H52 | Arkansas DPI | China QxIBV | |
| HT6 | 100% | 83% | 82% | 83% | 99% | 98% | 97% | 98% | 82% | 80% |
| HT9 | 83% | 100% | 85% | 99% | 83% | 83% | 83% | 83% | 82% | 84% |
| HT10 | 82% | 85% | 100% | 85% | 83% | 82% | 82% | 83% | 84% | 83% |
| HT11 | 83% | 99% | 85% | 100% | 83% | 83% | 83% | 83% | 82% | 84% |
The HT6 strain isolated from a poultry farm located in Ibagué showed closed relationship with the vaccine strain M41, with 99% of similarity. Between this strain and the H120, Conn 46, H52, Arkansas DPI and China QXIBV the identity was 98%, 97%, 98%, 82% and 80%, respectively. Among the isolates, the HT6 showed 83% of similarity with HT9 and HT11 and 82% with HT10. Two strains, one isolated on Coello (HT9) and the other in Ibagué (HT11) were grouped in the same cluster and were 99% similar, but these isolates were genetically distant from the reference strains. The HT10 strain isolated in Ibagué, showed a low similarity rate with the reference strains. Among the isolates, the HT10 showed 82% of similarity with the HT6 and 85% with HT9 and HT11.
Isolates of Colombia showed low similarity in nucleotide sequences with the Brazilian and Argentine isolates (<75%). However, they showed on average of 82% of similarity with the Cuban isolates (Table 3). HT6 showed 99% nucleotide identity with the isolated Cuba / La Habana / CB13 / 2009
DISCUSSION
The IBV is widespread in all the regions of the world causing economic losses to the poultry industry 1,8,9,11,22,23. Biosecurity practices associated with the control measures are used to prevent both the infection and the spread of IB on farms 3. One of the control measures are the vaccination protocols using both live attenuated and inactivated, but disease outbreaks continue to occur worldwide 5. In several regions of the world, including most of the South American countries, the Massachusetts is the only officially authorized strain for vaccine, but in some countries are released other strains such Connecticut, Arkansas, D207, D3896, 4/91 1,5. In Colombia, the use of vaccines containing serotype Massachusetts, Arkansas and Connecticut are allowed by Instituto Colombiano Agropecuário 6, but in the field are often used the vaccine containing the Massachusetts strain.
In this study, IBV field strains were isolated, same the poultry being vaccinated with Massachusetts serotype in the first days of life (Table 1). The HT6 strain showed 99% of similarity with a vaccine strain used in Colombia (M41). HT6 also showed hight similarity (99%) with Cuba/La Habana/CB13/2009 isolated. In the study of Acevedo et al 14, this isolate was grouped in the same clade of the M41 strain, in which HT6 also grouped.Therefore, it is believed that the HT6 strain may have been an isolated of vaccine strain. The isolation of vaccine strain in chickens with IB symptoms can be related to the virulence reversion of live attenuated vaccine which may cause dissemination and persistence of the vaccine virus 24, or even with the immunosuppression of birds with consequent increased vaccine reactions, resulting in clinical signs compatible with IB 10. In a study of phylogenetic relationship in Argentina, vaccine strains were also isolated in the country and they were grouped in the same cluster of the Massachusetts strains 10.
The other three strains were clustered with a genetic distance range from the vaccine strains, demonstrating that virus isolation in these poultry may have been a failure in vaccination programs. Poultry in the farms C and D were vaccinated in the 1st day of life whereas poultry from the farm B were vaccinated in the 10th day (Table 1). Several factors can influence the magnitude and the duration of the expected response, like the age of the poultry, maternal immunity levels, immunogenicity of the vaccine, interval between vaccination and infection and immune response of the host 5. It is believed that this case may have influenced by the use of a single vaccine dose in the first day of life, which is often insufficient to induce the necessary protection ranges throughout the life of the poultry. Besides that, the vaccine used containing serotype Massachusetts cannot have induced cross-protection against these genotypes 5. Although vaccines containing other strains are allowed in Colombia, as Arkansas and Connecticut, further studies should be performed to evaluate the range protection of these vaccines against these new genotypes, since the similarity presented was low, around 83%, among the isolates HT9, HT10 and HT11 and Massachusetts, Connecticut and Arkansas strains.
In a phylogenetic analysis study realized in Brazil, it was observed that the isolates were native and presented low identity with the vaccine strain. Moreover, they observed that the vaccine containing the Massachusetts strain unique freed by the Agricultural Defense Agency, probably did not supply cross-protection against these variants. As result, frequent outbreaks have been occurred 8. In another current phylogenetic study realized in Brazil it was observed that other IBV genotype had arisen and grouped separately from previous isolates in the country and also from the vaccine strain 11. This condition is not restricted to Brazil and it can be found in many other countries, such as Argentina 10, EEUU 25, Tunisia 9 and Australia 7. In these countries new genotypes also emerged. This shows the importance of molecular characterization of IBV in each region and the selection of strains which may be more suitable to the development of specific and effective vaccines against the disease.
The IBV is a RNA virus, and like other RNA virus, undergo frequent mutations in its genome due to the function of the errors done by RNA polymerase, as result causes deletions and mutation points 4,26. In addition, this virus has a high ability for genetic recombination (1, 4). These factors are involved in the outgrowth of new genotypes, with little change in the amino acids of the protein S1 already result in a change of serotype (4, 5, 24). Thus, genotyping of the IBV becomes an important tool for the epidemiologic study and identification of new genotypes, and also important to identify potential strains that could be used for the development of new vaccines 5.
The farms selected for the study housed poultry that presented respiratory symptoms, such as nasal discharge, snicking, sneezing, rales (the vibration emanating from lower in the respiratory tract watery), watery eyes and lethargy 2. The virus was isolated from trachea. The trachea is a major focus of the primary replication of IBV, reaching viral titer with maximum value in the first day post-infection 2. Despite the poultry presented respiratory signs, in most of the farms the IBV was not isolated, demonstrating that these signs can be confused with other respiratory diseases such as Laryngotracheitis and Coryza. Thus the diagnosis is essential for the confirmation of IB 27.
The HT9 and HT11 isolated of Ibagué and Coello cities, respectively, were grouped in the same cluster. On the other hand, these strains showed low similarity with the other isolates and reference strains. Already HT10 isolated in Ibagué showed low similarity with the other isolates and reference strains, being grouped separately for phylogenetic analysis. This demonstrates that new genotypes are present in Tolima Department, the most of them in Ibagué. This situation is critical, because in this case there is a risk of genetic recombination 1,4.
In conclusion there is an outgrowth of new genotypes in the Tolima Department, with a high risk of genetic recombination. The vaccine containing the Massachusetts strain was insufficient to induce protection against the circulating genotypes of IBV, demonstrating the necessity of using vaccines with different serotypes that induce cross-protection or develop new vaccines against these variants.
Conflicts of interest
The authors declare they have no conflicts of interest with regard to the work presented in this report.














