INTRODUCTION
Yeast of the genus Malassezia are found as commensal cutaneous microbiota in humans, as well as the external auditory canal in dogs. None the less, under humidity conditions, sweating, obstruction, keratinization disorders, and immunosupression, among others, they can cause diverse pathologies 1-4. In dogs the main diseases associated with these microorganisms are external otitis (EO) and dermatitis, with M. pachydermatis as the most frequent species reported. However, M. furfur has also been reported in patients with otic symptoms 5,6.
Prevalence of EO in dogs is between 10 - 20% (7-9), with its diagnosis based on clinical history or cytological findings (7,10). The gold standard assay is cell culture; however it is not a routine lab test. None the less, microbiological support is fundamental to define the etiology associated with these processes, as well as to contribute to epidemiology and clinical manifestations of this pathology.
In contrast to other species M. pachydermatis is a nonobligatory lipophylic. This yeast can grow in culture media devoid of lipids, such as Sabouraud's agar 1,11-13. Media utilized for obligatory lipophylic species isolation are modified Dixon agar and Leeming & Notman, containing Tween, glycerol, and oleic acid as lipid sources for yeast proliferation 11-12.
Malassezia spp. identification is performed by macroscopic morphological characterization of colony size, color, texture, surface, and margin. Microscopic measurements and biochemical and physiological findings confirm macroscopic findings. Among biochemical tests are catalase, β-glucosidase, proliferation on Sabouraud's agar without lipid supplements, Tween and Cremophor-EL assimilation, phospholipase activity, and pigment production in media with tryptophan as only source of nitrogen. Last, temperature dependant growth is also evaluated at 37°C and 40°C 2,12,13.
Other biochemical tests include positive urea reaction, which allows classifying yeasts of the genus Malassezia within the phylum Basidiomycota 14. Furthermore, catalase negative reaction permits to distinguish M. restricta from all other species within the genus 3,4,12. Other assays based on enzyme activity are phospholipase, hydrolyzing specific ester bonds in glycerophospholipids 15,16. In addition, tryptophan aminotransferase (TAM 1), in charge of tryptophan transamination yielding indol pyruvate; reacts spontaneously with remaining tryptophan molecules producing pigment 17. This enzyme has been widely studied by Zuther et al 18, in the fungus Ustilago maydis. In their work they demonstrated generated indolic pigments are identical to those produced by M. furfur, suggesting this enzyme could be involved in pigment production. Last, lipid assimilation assays evaluate yeast capacity to metabolize supplements of greater or lesser structural complexity such as Tween 20, 40, 60, 80 and Cremophor EL 3,12,13,19,20.
Moreover, physiological tests comprise temperature dependent growth, with 32°C being an optimal temperature. However, some species associated with dog skin pathologies can develop at greater temperatures. As a case in point, M. pachydermatis has the capacity to grow at 37°C and 40°C. In addition, M. furfur can proliferate at 41°C 12,21.
Taking into account EO in dogs has not established cell culture as a routine test, the objective of this work was to identify morphologically and biochemically Malassezia spp. from swab samples in dogs with EO.
MATERIALS AND METHODS
Microorganisms. From collected yeast samples stored at -80°C obtained from auditory canals in dogs diagnosed with EO from four veterinary clinics in Bogotá, 105 Malassezia isolates were characterized. All results were compared to reactions obtained from Centralbureau voor Schimmelcultures (CBS) reference strains: Malassezia furfur CBS 7019, Malassezia pachydermatis CBS 1879, Malassezia sympodialis CBS 7222, Malassezia slooffiae CBS 7956, and Malassezia globosa CBS 7966. Additionally, phospholipase positive control strains were Candida albicans ATCC 90028, Candida krusei ATCC 6258, and Candida parapsilosis ATCC 22019.
Morphological characterization. For morphological evaluation the isolates were thawed and reactivated in modified Dixon agar (two picks) and incubated at 32°C for 5 - 7 days. Macroscopic analysis was based on colony diameter measured in 10 isolated colonies from streaked dilutions. In addition, morphological characterization was performed according to color, texture, and margin. Microscopic evaluation included Gram stain performed from a fresh yeast suspension, to determine length and width (µm). Yeast without budding and with budding were included, for which the parent cell size was registered. Measurements in 30 yeast preparations were carried out by micrometry on both directions: length and width at 100 X 22.
Biochemical and physiological characterization. Biochemical and physiological tests were carried out to identify and characterize yeast from the genus Malassezia including enzymatic activities for catalase, urease, β-glucosidase, and phospholipase 2,3,12,15,16,19. In addition, pigment production in media with tryptophan as only source of nitrogen was also performed 23.
Except for M. pachydermatis, the remaining species in the Malassezia genus use lipids as a carbon source. Thus, lipid supplement assimilation in Cremophor EL (Sigma - USA) and different Tween 40 and 20 (Merck - Germany); Tween 60 and 80 (Sigma - USA) was used to define distinct species. Last, temperature dependant growth was also evaluated at 37°C and 40°C.
For phospholipase activity Sabouraud's agar was ammended with 10% egg yolk. The medium was inoculated from fresh cultures directly at four different points in the Petri dish, and incubated at 32°C for 21 days. To determine phospholipase activity the ratio of colony diameter to diameter of the dense white zone of precipitation around phospholipase positive colonies (Pz value) was used 15,16,24. No phospholipase activity was considered for a Pz value of 1 (Pz=1). High between 0.64 - 0.99, and very high for Pz values less than 0.64 15,24. Reading was performed after three weeks of culture averaging Pz values from the four different points of seeding.
For pigment production in media with tryptophan we employed Dixon agar with 0.6% L-trp. (Sigma - USA) 23. Petri plates were incubated at 32°C for 15 days, and the assay was positive if a brown diffusing stain was present on the media.
Statistical analysis. Data obtained from morphological analysis were analyzed with descriptive statistics taking into account central tendency and dispersion as a mean (X) and standard deviation (SD) and variation coefficient (VC), with values less than 15% considered acceptable 25, indicating data did not show large differences between them.
RESULTS
Morphological characterization. All morphological characteristics performed in this study are detailed in Table 1. Colonies with flat elevation, shiny and entire or undulate margins presented variability within the characterization, thus we were unable to group them according to pattern description.
Table 1 Canine Malassezia colonies macroscopic characterization.
| Species | Morphology variables | ||||
|---|---|---|---|---|---|
| CRS | CSR | CV | SM | VC | |
| Total isolates before identification | 43.80% (n=46) | 34.30% (n=36) | 21.90% (n=23) | 4.33 ±0.34 | 6.59 |
| M. pachydermatis (n=38) | 47.36% (n=18) | 28.95% (n=11) | 23.67% (n=9) | 4.90 ±0.28 | 6.85 |
| M. furfur (n=8) | 37.50% (n=3) | 37.50% (n=3) | 25.00% (n=2) | 4.80 ±0.28 | 6.41 |
| Malassezia spp. (n=59) | 42.37% (n=25) | 37.29 (n=22) | 20.33 (n=12) | 4.34 ±0.40 | 6.53 |
| CRS= Whitish colonies, rough surface, umbonate elevation, and undulated margins; CSR= Whitish colonies, slightly rough surface or smooth, umbonate elevation entire margins to slightly undulated; CV= Colonies with variable characteristics; VC=Variation Coefficient (%); SM= Size (mm) Mean±SD | |||||
Clinical isolate microscopic characterization did not demonstrate any difference among measurements with Gram stain and fresh yeast suspension, hence only Gram stain measurements are presented in Table 2.
Table 2 Cell morphometric characterization obtained from canine Malassezia isolates.
| Species | Parent cell Gram stain | |||
|---|---|---|---|---|
| LM | VC | WM | VC | |
| Total isolates before identification | 3.29±0.18 | 1.00 | 1.82±0.16 | 1.32 |
| M. pachydermatis (n=38) | 3.27±0.15 | 4.63 | 1.78±0.24 | 1.37 |
| M. furfur (n=8) | 3.33±0.28 | 8.47 | 1.88±0.25 | 1.05 |
| Malassezia spp. (n=59) | 3.29±0.18 | 5.38 | 1.83±0.25 | 1.37 |
| LM= Length (µm) Mean ± SD; VC=Variation coefficient (%); | WM= Width (µm) Mean ± SD | |||
Biochemical characterization. Positive urease activity for all CBS referenced strains and for 100% (n=105) isolates (Table 3) confirms their classification in the Basidiomycota phylum 14. Biochemical characterization is summarized in Table 3. Based on the biochemical profile 38 isolates corresponded to M. pachydermatis, 8 to M. furfur, and the remaining 59 could not be classified to the species level. Tables 1 - 3 describe morphological and biochemical characterization in detail for each species identified.
Table 3 Canine Malassezia isolates biochemical characterization.
| Biochemical test | Positive % (N=105) | M. pachydermatis (n= 38) | M. furfur (n= 8) | Malassezia spp. (n= 59) |
|---|---|---|---|---|
| Urease | 100 (n= 105) | n=38 | n=8 | n=59 |
| Catalase | 14.3 (n=15) | n=3 | n=3 | n=9 |
| Weak Catalase | 85.7 (n= 90) | n= 35 | n= 5 | n=50 |
| β-Glucosidase | 33.3 (n=35) | n=11 | n=3 | n=21 |
| Tween 20-40-60 y 80 assimilation | 19 (n=20) | n=6 | n=6 | n=8 |
| Tween 40, 60 y 80 assimilation | 51.4 (n=54) | n= 32 | n=2 | n=20 |
| Cremophor-EL assimilation | 34.3 (n=36) | n=4 | n=8 | n=24 |
| Sabouraud's agar growth | 70.5 (n=74) | n= 38 | _ | n=36 |
| Growth at 37°C | 100 (n=105) | n=38 | n=8 | n=59 |
| Growth at 40°C | 100 (n=105) | n=38 | n=8 | n=59 |
| Phospholipase activity | 66.7 (n= 69) High n=54 Very High n=15 | n=28 High: n=21 Very High n=7 | n=6 High: n=6 | n= 35 High: n=27 Very High n=8 |
| Pigment production in media with tryptophan | 40.95 (n=43) | Positive (n=8) Weak Positive (n=5) | Positive (n=8) | Positive (n=13) Weak Positive (n=9) |
| High Phospholipase activity: Pz 0.64-0.99. Very High Phospholipase activity: Pz <0.64 |
DISCUSSION
After test analysis for morphological, biochemical, and physiological characterization, 36.19% (n=38) isolates were identified as M. pachydermatis and 7.62 % (n=8) as M. furfur. The remaining 56.19% (n=59) isolates could not be identified at the species level, given the profiles did not coincide with those reported in the literature 11,14,19, nor with reactions presented for CBS reference strains.
Morphological, macroscopic, and microscopic characterization. According to macroscopic characterization based on reports by Torres et al 4, 78.3% (n=29) (Table 1) of isolates obtained from dogs in the clinic were identified as M. pachydermatis, they described opaque witish colonies, sometimes umbilicated with an average size of 4 - 5 mm 4. These results also agree with those reported by Guého-Kellermann et al 12 and Guého et al 13, who defined entire margins or slightly undulated. None the less, some differences were established with the reference strain M. pachydermatis CBS 1879, presenting opaque colonies, convex, witish with smooth surface with entire margins with an average size of 4 ± 1mm (VC = 6.45%).
In a first group identified as M. furfur, 37.5% (n=3) coincided with the morphology observed in M. furfur CBS 7019, with characteristics described by Guehó et al 12,13 for this species: witish colonies, smooth, with umbonate elevation, entire margins or slightly irregular with an average size of 5 mm 12. In contrast, the second group 37.5% (n=3) agrees with reports from Crespo et al 3, who described witish colonies, smooth or slightly rough with a 4-5 mm diameter 3. The remaining 25% presented variable macroscopic characteristics including flat elevation, undulate margins, corresponding partially to some reports 21.
Although 59 Malassezia spp., isolates shared some macroscopic features with M. furfur and/or M. pachydermatis, these were insufficient to characterize them up to species level, since biochemical tests presented atypical patterns for these two species 4,12.
Microscopic characterization of isolates identified as M. pachydermatis, evidenced oval cells and differences in measurements as compared to reports by Guillot et al 26, Dos Santos et al 21, and Guého-Kellermann et al 12, who described for this species oval cells with lengths between 4 - 5 µm and widths between 2 - 2.5 µm 12,21,26, as observed for the reference strain M. pachydermatis CBS 1879, with a mean cell length of 4.1 µm and 1.8 µm for cell width.
M. furfur isolates evidenced oval and cylindrical shaped cells of different sizes. This variability in shape concurs with pleomorphism reported for this species 3,12,21. Additionally, Crespo et al 3 reported average sizes of 4-6 µm. However, Dos Santos et al 21, and Guého-Kellermann et al 12 reported lengths up to 8 µm and widths of 1.5 - 3 µm, agreeing with the results here obtained. The average length for the reference strain M. furfur CBS 7019, was 5.48 µm and an average width of 2.0 µm.
Likewise, Malassezia spp., isolates microscopic characterization demonstrated attributes similar to those identified in M. furfur and M. pachydermatis in terms of morphological variability and cell size.
Colony measurement descriptive analysis suggests their sizes did not differ between species. Due to contrasting morphological characteristics present within species, morphometric parameters cannot be used as a guide to define distinct species within the Malassezia genus.
Biochemical and physiological characterization. According to biochemical characterization, catalase test was positive for all CBS reference strains and 100% isolates. This allowed inferring none of the clinical isolates was M. restricta3,4,12.
Out of the 38 clinical isolates identified as M. pachydermatis, only 11 were positive for bile esculine. This variability in β-glucosidase production has been reported for M. pachydermatis by Ashbee 2, Hossain et al 27, and Guého-Kellermann et al 12. This activity was also present for three M. furfur isolates, in agreement with reports from Mayser et al 19 and Guého-Kellermann et al 12, who describe M. furfur is capable of hydrolyzing esculine. Out of the remaining 59 Malassezia spp. samples, 21 were positive for bile esculine. This results confirm in this study esculine hydrolysis cannot be used to distinguish differences among species. In contrast, it can be used to distinguish between M. sympodialis and M. slooffiae, with the first one being positive for bile esculine 12.
The 105 evaluated isolates had different growth characteristics in presence of Cremophor EL and Tween 20, 40, 60, and 80, lipid supplements. According to Mayser et al 19, Cremophor EL is a test distinguishing M. furfur from other species. In this study, it was evidenced M. furfur CBS 7019 was the only reference species that had a positive reaction. None the less, Ashbee 2 and Guého-Kellermann et al 12 described variability in assimilation pattern for M. furfur and M. pachydermatis. In a study evaluating genetic variability, Hossain et al 27 confirmed Cremophor EL in 66% of the isolates studied. Based on these reports, the data here presented can be classified as M. pachydermatis and M. furfur.
With respect to Tween assimilation Ashbee 2 and Guého-Kellermann et al 12 proved M. pachydermatis is capable of Tween 20 assimilation, as was observed in 15.79% (n=6) of our isolates. However, 84.21% (n=32) and reference strain CBS 1879 did not assimilate Tween 20, as exemplified by Torres et al 4, and Crespo et al 3.
For M. furfur isolates, 75% (n=6) assimilated all Tweens, as reported by Crespo et al 3, Torres et al 4, and Guého-Kellermann et al 12. In addition, CBS 7019 reference strain was also capable of assimilating all Tweens. Never the less, 25% (n=2) assimilated only Tween 40, 60, and 80, a variation reported by Batra et al 28. Malassezia spp. exhibited different lipid supplement assimilation patterns. Therefore, they were grouped as follows: Group A (n=7) Tween 20, 60, and 80 assimilation; Group B (n=15) Tween 60 and 80 assimilation; Group C (n=9) due to increased variability it was not possible to classify. One of the isolates was unable to assimilate any Tween, thus it could correspond to M. globosa, however we did not observe the characteristic cerebriform colony morphology for this species.
The only species that did not present growth on Sabouraud's agar without lipid supplements was M. pachydermatis CBS 1879. This has been reported by different authors describing M. pachydermatis as the only nonbligatory lipohylic species within the genus 12,13. Likewise, clinical isolates compatible with M. pachydermatis grew on Sabouraud's agar. In addition, due to all Tween assimilation or Tween 40, 60, and 80 assimilation it was possibly to classify them as M. pachydermatis.
In regards to Malassezia spp. 61% could grow on Sabouraud's agar, classifying them as atypical M. pachydermatis12,13,26, although it should be taken into account that Cafarchia et al 29 reported lipodependent phenotypes for this species. In addition to other test results, such as Cremophor EL assimilation it was not possible to identify as M. pachydermatis with certainty.
Phospholipase activity was positive for all reference strains. Highest values were observed for M. furfur CBS 7019 (Pz=0.66) and M. pachydermatis CBS 1879 (Pz=0.62), in addition to all clinically isolated samples. Evidence of phospholipase activity coincides with results reported by Cafarchia et al 15 and Pini and Faggi 16, who illustrate this enzyme can be involved in pathogenesis, since it acts by hydrolyzing glycerophospholipid ester bonds. Phospholipids and proteins are important host's plasma membrane compounds. Thus, phospholipase activity induces arachidonic acid liberation, contributing to the inflammatory process through prostaglandin and leukotriene production, resulting in skin pH changes 12. Moreover, it contributes to host epithelial cell membrane disruption through pores formation, favoring tissue colonization 15. Results in this study evidenced M. pachydermatis had the highest phosphatase activity compared to other groups, in agreement with other reports in the literature 15,16.
All clinical isolates as well as M. furfur and M. furfur CBS 7019 reference strain were positive for pigment production in media with tryptophan, as the only nitrogen source. Data in this study corroborate results described by Lang et al 30. Colonies cultured in this media formed dark colonies with diffusible brown pigment after 10 - 12 days of incubation. Similar results have been reported in the literature, since these pigments allow differentiating M. furfur from other species in this genus. However, out of the 38 isolates characterized as M. pachydermatis, 13 produced pigment, five had a faint brown pigmentation in the middle, and eight produced pigment at a different intensity (Figure 1). Recently, it has been reported some M. pachydermatis strains produce pigment 27. Mayser et al 23, evaluated pigment production in M. furfur, M. pachydermatis, and M. sympodialis in different culture media: minimum medium (p-medium) and Dixon agar supplemented with 0.6% L-trp. All M. furfur isolates produced pigment in both media; on the contrary, only one of the M. pachydermatis isolates produced slight pigment in Dixon agar with 0.6% L- trp. Furthermore, in a study carried out by Hossain et al 27, it was observed that 84 M. pachydermatis isolates out of 210 evaluated, produced pigments at variable intensities. In addition, pigment production can be associated with strain genetic variations 26, as observed in this study.
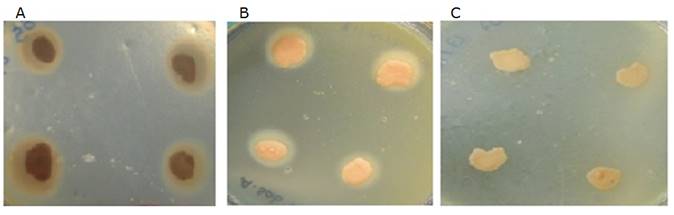
Figure 1 Canine Malassezia phospholipase activity. A Very high activity. B High activity. C No activity.
Under physiological conditions M. pachydermatis and M. furfur can proliferate at 37°C and 40°C 13,26. These results permitted to discriminate the presence of species such as M. obtusa, M. restricta, and M. globosa uncapable of growing at 40°C 12.
In conclusion, morphological and biochemical characteristics permitted to identify 36.19% (n=38) of all isolates as M. pachydermatis and 7.62% (n=8) as M. furfur. However, 56.19% (n=59) could not be defined as a specific species. Although it was feasible to classify them within the Malassezia genus based on positive urease activity and morphology; the atypical phenotypical traits of isolates reported as Malassezia spp. could correspond to Malassezia furfur or M. pachydermatis variants. Therefore, a polyphasic strategy incorporating molecular biology tools is required to identify variants within the Malassezia genus.
Conflict of interest
The authors report no conflict of interest.














