INTRODUCTION
Antimicrobial resistance is a serious threat to global public health 1 because antimicrobial use is an important factor in maintaining human and animal health worldwide 2. Antimicrobial-resistant populations are present in all bacterial communities 3, thus representing a serious threat to both human and animal populations, especially considering the potential transfer of resistance and zoonotic diseases via the food chain to humans 2,4. The main causes of antimicrobial resistance in food animals seem mainly to be excessive use of antimicrobials, incorrect antimicrobial dosage and unregulated access to drugs 5. Therefore, and considering that antimicrobial agents are the most important therapeutic tool against bacterial diseases in both human and animals 5,6, it is of the upmost relevance to avoid development of antimicrobial resistance in bacteria to ensure therapeutic efficacy. Therefore, prudent use of antimicrobials is necessary 7 combined with overall coordination of medication, governmental regulation and surveillance 5.
Few studies have been done in Chile regarding antimicrobial resistance in cattle - most antimicrobial resistance studies in Chile have been performed in animals such as poultry and swine 8,9. Previous work in dairy farms and cattle - performed more than two decades ago - focused on subclinical and clinical mastitis in both central and southern Chile followed by work on mastitis and antimicrobial resistance 10,11. Additional research concentrated mainly on cattle in the central region looking at Escherichia coli from fecal samples in dairy and beef cattle 12. Globally, some studies have described antimicrobial resistance trends for E. coli in multiple species including calves 13,14, although work tends to focus on specific genetic characterization of isolates 8,15. These studies of antimicrobial resistance often have shown resistance patterns that are highly variable among and within farms.
Antimicrobial resistance monitoring programs have been established in several countries, like the Danish Integrated Antimicrobial Resistance Monitoring and Research Program (DANMAP) 16 or the United States of America National Antimicrobial Resistance Monitoring System (NARMS) 6 among others. Chile recently started its “National Antimicrobial Resistance Control Plan”. It is officially required a prescription for purchasing antimicrobials. Even though it is mandatory, the authors have observed that it is a common practice to sell antimicrobials without prescription.
In some developing countries, it is believed that the unrestricted use of antimicrobials could be producing widespread resistance 17, although little information is available. Some antimicrobials are being described by clinicians as useless for control of diarrhea and pneumonia in calves or mastitis in dairy cows. This could be explained by incorrect diagnosis, inappropriate treatment (e.g., dosage) or to antimicrobial resistance.
This study, based on historical records from a university reference diagnostic laboratory, aims to identify phenotypic antimicrobial resistance findings in calves, in association to antimicrobial use, antimicrobial resistance patterns and practical association to the effectiveness of antimicrobials in the field.
MATERIALS AND METHODS
Study location. Information was collected from necropsy records from 2002 through 2015 from the Veterinary Anatomic Pathology Laboratory at Universidad Austral de Chile. (In the years 2008 and 2009 no antimicrobial resistance evaluations were done due to lack of infrastructure after a fire destroyed the microbiology laboratories). This Laboratory is located in the Los Ríos region in southern Chile, in the city of Valdivia. The coordinates are: latitude -39.804437, longitude -73.252787 and altitude 5 m.
Records. Records of all calves <30 days old with antimicrobial resistance test performed on bacterial isolates were selected for study. Information collected included animal information like sex, age, breed and production use (beef or dairy). Basic farm information included owner name, address, commune and region. Temporal information like date of reception of the animal/sample, date of necropsy and antimicrobial resistance results was also retrieved from the records.
Data analysis. Based on the evaluation of 17 commonly used antimicrobials, isolates were classified as either resistant (including non-conclusive results) or sensitive to antimicrobials. Each isolate was tested for a variety of antimicrobials, ranging from four to eleven (Table 1). When receiving the sample, the laboratory collected information on whether the submitted animal had been treated with antimicrobials (type & dose not specified). Overall results were recorded in an MS Excel© database. Isolates considered in this study include only E. coli, β-hemolytic E. coli and Salmonella spp. Most calves were submitted after serious negative health events occurred, associated mortalities, diarrhea and respiratory disease events in calf pens (data not shown).
Table 1 Antimicrobial resistance score (AMR score*) from isolates obtained from <30-day-old calves between 2002 and 2015 in southern Chile.
| Isolate | AMR score | ||||
|---|---|---|---|---|---|
| No. | Average | S.D. | Min | Max | |
| E. coli | 61 | 1.75 | 0.5 | 1 | 2.7 |
| β-hemolytic E. coli | 27 | 1.67 | 0.5 | 1 | 3.0 |
| Salmonella spp. | 19 | 1.72 | 0.6 | 1 | 2.5 |
| * An isolate identified as sensitive to an antimicrobial was assigned a score of “1”. A non-conclusive or a resistant isolate received an AMR score of “3”. AMR scores were added and averaged per sample. If the final averaged score was 1, the sample was considered “fully sensitive” and if the score was >1, the sample was considered “resistant”. An AMR score of “3” means resistance to all antimicrobials tested. | |||||
Classical microbiology procedures were followed for the isolation and identification of E. coli and Salmonella spp. from either feces or tissue smears, using CLSI standard procedures 18. All isolates were tested against a different set of antimicrobials using disc diffusion method 18. The set of antimicrobials to be tested depended on recommendations from the field veterinarian, pathologist or the laboratory personnel, and therefore there was large variation in the number of antimicrobials tested on each isolate, ranging from four to eleven. Overall 17 different antimicrobials were tested: gentamicin, ampicillin, amoxicillin, cefacetrile, cefapirin, cefuroxime, cefoperazone, ceftiofur, ceftriaxone, cefquinome, florfenicol, danofloxacin, enrofloxacin, nalidixic acid, doxycycline, oxytetracycline and trimethoprim/sulfamethoxazole.
Antimicrobial resistance score. Basic data description was performed on the antimicrobial resistance cases identified. Antimicrobial resistance was described for each isolate species, in association with the number of reported antimicrobials used and per year throughout the study period. Antimicrobial resistance findings where coded into a score (antimicrobial resistance score = AMR score), to aggregate all the available resistance information (E. coli and Salmonella spp. isolates, antimicrobials, years, etc.) to identify overall resistance pattern. An isolate identified as sensitive to an antimicrobial was assigned a score of “1”. A non-conclusive or a resistant isolate received an AMR score of “3”. AMR scores were added and averaged per sample.
- If the final averaged score was 1, the sample was considered “fully sensitive”
- If the score was >1, the sample was considered “resistant”. With the aggregated data that included information on all bacterial species isolated and antimicrobials tested, an overall resistance pattern was constructed.
Additionally, and based on the antimicrobial resistance classifications published by Magiorakos (19), all isolates were classified in either “Sensitive” (low or no resistance found), MDR - multi drug resistant: non-susceptible to ≥1 agent in ≥3 antimicrobial categories, Possible XDR (extensively drug resistant): non-susceptible to ≥1 agent in all but ≥2 categories and Possible PDR (pan drug resistant): non-susceptible to all antimicrobial agents listed. Further comparisons for AMR patterns were performed using classic statistical methods for non-normally distributed data, like the Kruskal-Wallis test (p=0.05).
RESULTS
Overall 107 antimicrobial resistance evaluations were included in this study. Most animals originated in the Los Ríos and neighboring regions: 55 (51%) Los Ríos, 43 (40%) Los Lagos. Nine calves (8%) came from the Araucanía, Bíobío and unrecorded regions. All these regions are in central southern Chile, roughly between 36-43° South and 74-72° West. Also, 93 (87%) samples were from dairy calves and 15 (14%) from beef or dual production calves. The main breed reported was Frisón Negro (53 animals, 50%) followed by 30 (28%) Holstein-Friesian animals. Other reported breeds (26%) included local breeds like Overo Colorado, Angus and Wagyu calves. Reported sex of the animals was 53 (50%) males and 45 (42%) females, with 11 (10%) missing values. Fifty percent of the calves included in this study were <10 days old and 50% between 10 and 29 days old.
Of the collected samples, 29 (27%) came from animals already treated with antimicrobials, 36 (34%) samples were from untreated calves and 44 (41%) of records did not contain this information. Sixty-one of the isolates were E. coli, 27 were β-hemolytic E. coli and 19 Salmonella spp. The antimicrobial resistance score (AMR) was calculated for each of the isolates. This allowed statistical description of overall antimicrobial resistance patterns for E. coli, β-hemolytic E. coli and Salmonella spp. (Table 1). AMR scores were similar among the 3 bacterial groups, with no statistically significant differences found among groups (Kruskal-Wallis, p=0.79).
The yearly overall antimicrobial resistance data can be observed in figure 1. When observing the changes in AMR score in relation in calve age, it was seen that the averages score remains practically the same, with no greater resistance found in older calves and no statistical differences among age groups (Kruskal-Wallis test, p=0.97; data not shown). Isolates were classified as “sensitive” and “multi resistant” (MDR + possible XDR + possible PDR) (Figure 2) and a more detailed classification in “sensitive”, “MDR”, “possible XDR” and “possible PDR” can be observed in table 2. AMR scores for antimicrobials tested 10 or more times are listed in table 3. E. coli was highly susceptible to ceftriaxone (AMR score = 1.0) and ceftiofur & cefoperazone (AMR score = 1.1); β-hemolytic E. coli to florfenicol, ceftiofur and gentamicin (AMR score = 1.4); and Salmonella spp. to gentamicin (AMR score = 1.0).
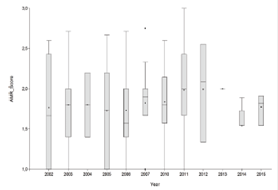
Figure 1 Overall antimicrobial resistance score (AMR score) from E. coli, β-hemolytic E. coli and Salmonella spp. isolates obtained from up <30 day old calves between 2002 and 2015. *No data available for 2008 and 2009. An isolate identified as sensitive to an antimicrobial was assigned a score of “1”. A non-conclusive or a resistant isolate received an AMR score of “3”. AMR scores were added and averaged per sample. If the final averaged score was 1, the simple was considered “fully sensitive” and if the score was >1, the sample was considered “resistant”. Boxes represent data within the 0.25 and 0.75 quantiles. Whiskers, the 0.05 and 0.95 quantiles.
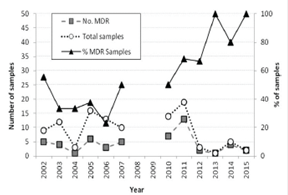
Figure 2 Yearly antimicrobial resistance findings from E. coli, β-hemolytic E. coli and Salmonella spp. isolates obtained from <30 day old calves between 2002 and 2015. All isolates were classified in either “Sensitive” (low or no resistance found) or MDR - multi drug resistant: non-susceptible to at least ≥1 agent in ≥3 antimicrobial categories
Table 2 Antimicrobial resistance findings from E. coli, β-hemolytic E. coli and Salmonella spp.isolates obtained from <30 day old calves between 2002 and 2015.
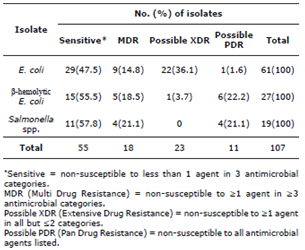
Table 3 Antimicrobial resistance findings from E. coli, β-hemolytic E. coli and Salmonella spp. isolates obtained from <30 day old calves between 2002 and 2015. All antimicrobials that were tested at least 10 times are included. n represents the number of isolates tested per antimicrobial agent.
| E. coli | AMR* score | n | |
|---|---|---|---|
| Ceftriaxone | 1.0 | 12 | |
| Ceftiofur | 1.1 | 19 | |
| Cefoperazone | 1.1 | 14 | |
| Cefquinome | 1.4 | 16 | |
| Danofloxacin | 1.4 | 25 | |
| Florfenicol | 1.6 | 55 | |
| Gentamicin | 1.7 | 49 | |
| Enrofloxacin | 1.8 | 41 | |
| Cefuroxime | 1.9 | 22 | |
| Nalidixic acid | 2.1 | 18 | |
| Trimethoprim/sulfamethoxazole | 2.3 | 57 | |
| Oxytetracycline | 2.6 | 43 | |
| Amoxicillin | 2.9 | 20 | |
| E. coli β-hemolytic | AMR* score | n | |
| Florfenicol | 1.4 | 27 | |
| Ceftiofur | 1.4 | 16 | |
| Gentamicin | 1.4 | 23 | |
| Danofloxacin | 1.6 | 18 | |
| Enrofloxacin | 1.6 | 25 | |
| Oxytetracycline | 2.2 | 17 | |
| Trimethoprim/sulfamethoxazole | 2.5 | 20 | |
| Ampicillin | 2.7 | 14 | |
| Salmonella spp. | AMR* score | n | |
| Gentamicin | 1.0 | 16 | |
| Danofloxacin | 1.4 | 12 | |
| Trimethoprim/sulfamethoxazole | 1.8 | 19 | |
| Enrofloxacin | 1.8 | 12 | |
| Florfenicol | 1.8 | 18 | |
| Amoxicillin | 2.2 | 12 | |
| Oxytetracycline | 2.3 | 14 | |
| *Antimicrobial resistance results in 3 points and one sensitive result is 1 point for each specific drug. These are averaged by the number of test performed on each sample to produce the Antimicrobial Resistance Score. A drug with and AMR Score of 1 resulted in 100% sensitivity in all isolates tested. A score of 3, 100% resistance. |
From all isolates studied, the antimicrobial resistance pattern was summarized in table 4. The most common multidrug resistance phenotype observed in E. coli was amoxicillin, cefuroxime, florfenicol, enrofloxacin, oxytetracycline and trimethoprim/sulfamethoxazole (in 5 isolates); for β-hemolytic E-coli this phenotype was oxytetracycline and trimethoprim/sulfamethoxazole (in 4 isolates); and for Salmonella spp. it was florfenicol, trimethoprim/sulfamethoxazole and amoxicillin, cefuroxime and oxytetracycline with 3 isolates each. No statistically significant differences were found (Kruskal-Wallis p=0.27) between treated, unknown treatment and untreated animals, with 20% of missing values.
Table 4 Antimicrobial resistance patterns for E. coli, β-hemolytic E. coli and Salmonella spp. isolates obtained from <30 day old calves between 2002 and 2015.
| Antimicrobial resistance patterns* | No. (%) of isolates | |||||
|---|---|---|---|---|---|---|
| E. coli (n=61) | β-Hem E. coli (n=27) | Salmonella spp. (n=19) | ||||
| All sensitive | 6 | (10) | 5 | (19) | 6 | (32) |
| AMP | 1 | (2) | ||||
| GEN | 3 | (5) | ||||
| OXT | 4 | (7) | 1 | (4) | ||
| SXT | 1 | (2) | 1 | (5) | ||
| AMP, CEF | 2 | (7) | ||||
| AMP, ENR | 1 | (4) | ||||
| AMP, SXT | 1 | (4) | ||||
| AMX, OXT | 3 | (5) | ||||
| FLO, SXT | 3 | (16) | ||||
| GEN, OXT | 2 | (3) | ||||
| GEN, SXT | 2 | (3) | ||||
| NAC, OXT | 1 | (2) | ||||
| NAC, SXT | 2 | (3) | ||||
| OXT, SXT | 4 | (7) | 4 | (15) | 1 | (5) |
| AMP, OXT, SXT | 1 | (4) | ||||
| AMX, CEF, OXT | 2 | (3) | 2 | (11) | ||
| AMX, OXT, SXT | 1 | (2) | ||||
| CFQ, NAC, SXT | 2 | (3) | ||||
| GEN, AMP, CEF | 1 | (4) | ||||
| GEN, AMP, SXT | 1 | (2) | ||||
| GEN, CFC, SXT | 2 | (7) | ||||
| GEN, FLO, NAC | 1 | (2) | ||||
| GEN, FLO, SXT | 1 | (2) | ||||
| GEN, OXT, SXT | 2 | (3) | ||||
| AMP, CFC, OXT, SXT | 2 | (7) | ||||
| AMP, ENR, OXT, SXT | 1 | (4) | ||||
| AMX, CEF, ENR, OXT | 1 | (2) | 1 | (5) | ||
| AMX, CEF, OXT, SXT | 1 | (2) | ||||
| AMX, DAN, ENR, OXT | 1 | (2) | ||||
| AMX, ENR, OXT, SXT | 1 | (2) | ||||
| CEF, ENR, NAC, SXT | 1 | (2) | ||||
| CEF, FLO, DAN, ENR | 1 | (4) | ||||
| CEP, FLO, DOX, SXT | 1 | (4) | ||||
| FLO, DAN, NAC, SXT | 1 | (2) | ||||
| FLO, ENR, NAC, OXT | 1 | (5) | ||||
| FLO, ENR, NAC, SXT | 1 | (2) | ||||
| GEN, AMP, OXT, SXT | 1 | (2) | ||||
| GEN, FLO, ENR, SXT | 3 | (5) | ||||
| AMP, AMX, DAN, ENR, SXT | 1 | (4) | ||||
| AMP, FLO, DAN, ENR, SXT | 1 | (4) | ||||
| AMX, CEF, FLO, ENR, OXT | 1 | (5) | ||||
| AMX, CEF, FLO, OXT, SXT | 1 | (5) | ||||
| AMX, FLO, ENR, OXT, SXT | 3 | (5) | ||||
| CFQ, DAN, NAC, OXT, SXT | 1 | (2) | ||||
| AMX, CEF, FLO, ENR, OXT, SXT | 5 | (8) | ||||
| AMX, FLO, DAN, ENR, OXT, SXT | 2 | (11) | ||||
| GEN, AMX, FLO, DAN, OXT, SXT | 1 | (2) | ||||
| GEN, CEF, CEZ, FLO, DAN, ENR, SXT | 1 | (2) | ||||
| GEN, FLO, DAN, ENR, NAC, OXT, SXT | 1 | (4) | ||||
| GEN, AMP, AMX, FLO, DAN, ENR, DOX | 1 | (4) | ||||
| *AMP=ampicillin, GEN=gentamicin, OXT=oxytetracyclin, SXT=trimethoprim/sulfamethoxazole, CEF=cefuroxime, ENR=enrofloxacin, AMX=amoxicilin, CFQ=cefquinome, NAC=nalidixic acid, FLO=florfenicol, CFC=cefacetrile, DAN=danofloxacine, CEZ=cefuroxime, DOX=doxyciclin. | ||||||
DISCUSSION
Antimicrobial resistance is a major threat to human and animal health worldwide 13. Although it involves multiple animal species and multiple bacteria in very complex associations, studies typically address the topic a single species, single agent or single environment issue rather than the global situation 20,21. This work, although focused in dairy and beef calves in a particular region, proposes a new approach to antimicrobial resistance evaluation, allowing for easy visualization of global trends in multiple bacterial isolates for more than a decade of data (in this case, in E. coli and Salmonella spp.). This is important if we consider that resistance genes can be transferred among bacteria of different species in the environment 14. As observed in figure 1, the general antimicrobial resistance yearly scores do not show an evident tendency, with no steady increase or decrease of AMR yearly scores. Nevertheless, during the last study years 2011-2015 an increase in the yearly AMR score was observed. However, the number of isolates studied decreased, resulting in an increased variability of the sample. When summarizing the data in MDR (Figure 2), it can be observed that there was an increase in the percentage of MDR samples starting in 2011, although, as mentioned, the number of samples analyzed per year decreased.
Antimicrobial resistance patterns, as described in Table 4, show an important number of resistant isolates, with a very large variation on the phenotypic resistance patterns, that include resistance to most antimicrobial families. Samples show resistance to up to 7 different antimicrobials in 2 isolates. In our study, 76% of E. coli isolates showed resistance to 2 or more antimicrobials and 27% of E. coli isolates showed resistance to 4 or more antimicrobials. This is similar to results obtained in a large retrospective study in the US 21 where 71% of E. coli isolates showed resistance to 2 or more antimicrobials, and with results from Belgium 13, where an average resistance against at least one antimicrobial was found in 82.14% of E. coli isolates, similar to the 90% and 81% found in E. coli and β-hemolytic E. coli respectively in this study (Table 4).
Our findings are consistent with an environment where bacteria have been exposed to multiple drugs, quite possibly involving incorrect dosage and treatment periods. In this regard, a report on sub therapeutic treatments indicated that AMR treatments had only a limited impact on the nature of antimicrobial resistance in E. coli, most commonly describing resistance to tetracycline, sulfamethoxazole, ampicillin, chloramphenicol and streptomycin 22. On the other hand, our results contrast largely with findings from adult cattle at slaughter where over 90% of E. coli and Salmonella spp. samples showed no resistance at all 23. We found only 10% of E. coli, 19% of β-hemolytic E. coli and 29% of Salmonella spp. isolates sensitive to all antimicrobials tested, results similar results to those previously reported, where 11.7-18.2 to 39% of E. coli isolates were all sensitive E. coli24,25. Isolates in our study originated from sick calves, some of which had been subject of unsuccessful antimicrobial treatments. Moreover, all calves originated from premises with serious ongoing health events. Traditionally, calves are only submitted to a laboratory when treatments fail. In this context, it is interesting to observe that isolates from calves reported as “not treated” did not tend to be less resistant than those of calves reported as treated. These results are similar to findings in Canadian calves, where no association between previous treatment and AMR was found 26. This can be attributed either to poor records (in association to recall bias or failure to provide or register accurate information) or to the possibility that the untreated calves acquire highly resistant agents from the environment without having received treatment themselves.
We found no association between the calf age and AMR findings. These results agree with a study that observed no effect of age on antimicrobial resistance in calves, while evaluating the effect on housing type and resistance 25. In contrast, other studies reported an effect of age in AMR 14,26. Nevertheless, it has been reported that ampicillin-resistant E. coli levels were typically low in the first 8 months of age 27, and our specimens were from calves <30 days of age.
Multi-resistant strains in very young calves was found, perhaps indicating that isolates had insufficient time to mutate in the host. These multi-resistant bacteria could have originated from the environment and likely reflect a complex exchange, genetic mixing and relationships among bacterial populations, hosts and the environment 28,29. These largely unknown processes could result in the finding of highly resistant bacteria in young calves, no increase in resistance with age (or even a decrease) and, no clearly defined trend at the yearly level. Furthermore, no statistical association was found between calf sex and AMR score, as previously reported 26.
The main limitations of this study were that samples submitted were a convenience sample of farms and therefore not representative of a larger population. Nevertheless, these results may be a good estimation of the situation regarding AMR in farms under treatment in normal field conditions, sometimes with poor veterinary supervision, no antimicrobial resistance evaluation, prior treatment and poor antibiotic administration practices and inaccurate dosage. Also, as in most retrospective studies, data records were not necessarily complete or accurate. Records were collected for practical purposes and future research is normally not considered as relevant at the time of collection. Finally, the number of samples received per year was highly variable and, for reasons not clear to the authors, very few isolates were received between 2013 and 2015.
At the practical level, it is important to note that this retrospective study allowed researchers to describe available data to provide useful information for veterinary practitioners and farmers. Based on our results, it is evident that antimicrobials like oxytetracycline and amoxicillin should be avoided when treating suspected E. coli diarrhea cases, and that ampicillin and trimethoprim/sulfamethoxazole are ineffective alternatives. These findings are similar to extensive AMR to tetracycline and trimethoprim/sulfamethoxazole found in Canada in 2008 30. Also, in cases of suspected Salmonella spp. outbreaks in calves, amoxicillin and oxytetracycline should be avoided due to extensive resistance. These results are similar to what was found in central New York, where ampicillin was also found as a prevalent AMR phenotype among other resistances 25. Ceftiofur should be the recommended treatment for E. coli diarrheas, while gentamicin appears to be the recommended antimicrobial for Salmonella spp. outbreaks, in the lack of an antimicrobial resistance evaluation. It is relevant to note that not a single isolate showed resistance to ceftriaxone, a drug that is not approved in Chile for veterinary use and therefore the exposure of these bacterial field strains was expected to be close to 0. This drug could work as an AMR control group, indicating the expected level of AMR to an antimicrobial not available in the market.














