INTRODUCTION
The sheep production systems in Colombia are based on the Colombian Creole sheep breed (OPC; by its acronyms in Spanish). The OPC presents important adaptive characteristics to the tropical climate, such as heat tolerance, ectoparasites and the ability to consume pastures of low nutritional value [1]. However, these systems have been considered as a secondary activity of livestock that is carried out in traditional and/or family systems, with low input requirements and in mixed production systems with other species [2,3], which entails to which problems in nutrition, reproduction, health, genetic progress, animal welfare, and business management; among others, are common and are reflected in low productive efficiency [1,4].
There are few breeding programs in the OPC, based on absorbent crosses with imported breeds to take advantage of the hybrid vigor [5]. However, in order to increase the competitiveness of the sector, it is necessary to incorporate new technologies or management practices that generate competitive advantages through knowledge management systems that adapt to the conditions of primary production in a developing country like Colombia [2,4,6]. This is how molecular tools are used in animal genetic improvement programs, through molecular marker-assisted selection. This practice improves accuracy and increases genetic progress, through the identification, mapping and analysis of polymorphism of the genes involved in the main metabolic pathways related to animal growth, the distribution of nutrients in different tissues and product quality [7]. Calpain (CAPN) and calpastatin (CAST) can be considered as candidate genes with the potential to be used in selection programs since single nucleotide polymorphisms (SNPs) have been associated with improvement in meat quality [8].
The CAPN gene is associated with the degradation of postmortem myofibrillar proteins, causing softening of the meat; is located on chromosome seven and has 25 exons. The SPN g.24962426T> C located in exon ten has been the more studied [9]. On the other hand, the CAST gene is the main inhibitor of the action of calpain slowing down the speed and the degree of postmortem softening of the meat [8]. It is located in chromosome 5 of the sheep, the calpains system is made up of the genes of u-calpain (CAPN1), m-calpain (CAPN2) and calpain 3 (CAPN3), in which numerous SNPs correlated with gene expression and meat tenderness have been identified in different species [9,10]. Also, the calpains are involved in muscle growth at different stages of development and in the degradation of muscle proteins, influencing the loss of muscle mass [7,8,9]."abstract":"Calpastatin (CAST
Reports of studies of polymorphisms in the afore-mentioned genes in sheep from Colombia are limited. Therefore, the objective of this work was to characterize the genetic polymorphism type SNPs in the calpain and calpastatina genes in the OPC breed.
MATERIALS AND METHODS
Populations, blood collection, and DNA extraction. In the present investigation, 300 individuals belonging to two OPC subpopulations located in the departments of Valle del Cauca (VC, n = 150) and Sucre (SC, n = 150) were used. Blood samples were collected in tubes with anticoagulant (EDTA 7.2 mg) considering the procedures of sample collection, management, and conservation, the ethical, technical, scientific and administrative standards for animal research contained in Law 84 (National Congress of Colombia, 1989). DNA extraction was performed using the QIAamp® DNA Mini Kit from QIAGEN. The quantity and quality of the DNA were evaluated using NanoDrop 2000TM (Thermo Fisher Scientific).
Amplification and genotyping of the CAPN and CAST loci. All PCR reactions were carried out in a final volume of 25 μl containing 20 ng of DNA, 250 nM of each primer and 1X of the super mix MangoMix™ (Bioline©). The primers used for the CAPN locus (CAPNS1, accession: XM_027514355.1) were 5'-AACATTCTCAACAAAGTGGTG-3' and 5'-ACATCCATTACAGCCACCAT-3' [11], for the CAST locus (CAST-A, accession: KX_722534.1) 5'-TGGGGCCCAATGACGCCATCGATG-3 'and 5 '-GGTGGAGCAGCACTTCTGATCACC-3' [12]. The thermal profiles were initially denatured at 95°C for 5 minutes. Followed by 35 cycles of 95°C for 30s, 60°C for 60s and 72°C for 60s for CAPN and 95°C for 60s, 58°C for 60s, 72°C for 60s for CAST; followed by a final extension of 10 minutes at 72°C. The amplifications were performed in a MasterCycler Nexus Gradient thermocycler from Eppendorf®. Visualization of the amplified fragments was verified by electrophoresis in 1.2% agarose gels at 80 V for 45 minutes stained with GelRed™ (Biotium, Inc. USA).
The genotypes for the SNPs studied in the CAST locus were obtained by PCR-RFLP with the restriction enzyme MspI, in a final volume of 15 μl containing in 5 μl of the PCR product, 1U of the MspI enzyme and 1X of the tampon. The incubation was at 37°C for two hours, followed by 80°C for 20 minutes. The digestion products were visualized in 9% polyacrylamide gels (Acrylamide: Bis-acrylamide 37:1) at 150 V for 40 minutes and stained with GelRed™ (Biotium, Inc. USA). Genotypes were identified as reported by Azari et al [11] and Santos et al [12].
For the CAPN locus, the genotypes were obtained by PCR-SSCP, in 10% polyacrylamide-denaturing gels (100:1, Acrylamide: Bis-acrylamide) and 3.7% glycerol. Each sample was denatured with formamide (95% formamide, 0.05% Xylene-Cyanol, 0.05% bromophenol blue, 20 mM EDTA pH 8.0) and subjected to 95 ° C for 5 minutes and then cooled to 4°C. The gels were run at 190 volts for 5.5 hours, in a Multigel Biometra chamber. The staining was performed with GelRed™ (Biotium). The genotypes were identified as reported by Azari et al [11].
Data analysis. The genotypic and allelic frequencies, the observed (Ho) and expected heterozygosity (He) and the fixation index (F) for each subpopulation and in total were calculated. The deviations from the Hardy-Weinberg equilibrium (EHW) were estimated and the molecular analysis of variance was performed to estimate the values of FST, FIS, and FIT. The genotypic and allelic frequencies found between loci and subpopulation were compared using the Fisher test with a significance of 5%. All analyzes were performed with the Arlequin programs ver 3.5.2.2 [13] and GENALEX ver 6.5 [14].
RESULTS
The two SNPs evaluated were polymorphic. The allelic and genotypic frequencies did not differ significantly (p> 0.05) between subpopulations for both loci.
In the CAST locus, for the whole population, the MM genotype was the most frequent, followed by the other genotypes (Table 1). The MM genotype was more frequent in the SC subpopulation than in the VC subpopulation, although the opposite occurred in the MN genotype. While the NN genotype presented the same frequency in both subpopulations (6%). Regarding the allelic frequencies, the M allele exceeded the N allele in frequency (Table 1). The M allele was more frequent in the SC subpopulation and the N allele was the most frequent in the VC subpopulation.
Table 1 Genotypic and allelic frequencies at the CAST and CAPN loci in the OPC breed.
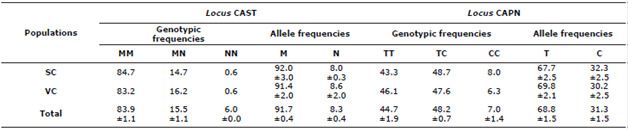
SC: Sucre. VC: Valle del Cauca.
For the CAPN locus, a greater frequency of the heterozygous genotype was found in the whole population (48.2±0.7%) and in the subpopulations SC (48.7%) and VC (47.6%). The CC genotype had the lowest frequency (Table 1). The T allele of the CAPN locus presented a frequency of 68.8 ± 1.5% for the entire population, being higher in the VC subpopulation, on the other hand, the C allele presented higher frequency in the SC subpopulation (Table 1).
Table 2 shows the values of genetic diversity in the OPC population studied. In the CAST locus, the values of Ho and He averages were similar with a slight excess of heterozygotes not significant; on the other hand, in the CAPN locus this excess induced a negative value of F and significant deviations of the EHW (p<0.05). The Ho and He values for the CAST locus were higher in the VC subpopulation and in the CAPN locus in the SC subpopulation. Only the value of F was positive in the subpopulation SC for the CAST locus (0.004) the other values were negative. The locus-to-locus analysis indicated that all were in EHW.
Table 2 Ho, He, F values and deviations of the EHW at the loci studied in the OPC breed.
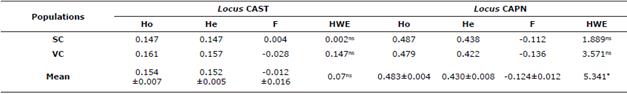
SC: Sucre. VC: Valle del Cauca. Ho: observed heterozygosity. He: expected heterozygosity. F: fixation index. HWE: Hardy-Weinberg equilibrium (qui-square value). *p<0.05.
The analysis of molecular variance showed that 100% of the variation is due to differences within individuals, with non-significant values (p>0.05) of FST, FIS and FIT of 0.002, -0.093 and -0.095, respectively.
DISCUSSION
In the CAST locus (AF016006, AF016007, and AF016008.1) fragments digested by the MspI enzyme were found between exons 1C and 1D, where a nucleotide substitution of A by G occurs [15]. In Dalagh sheep genotypes MM (36%), MN (38%) and NN (26%) were found with higher frequency of the M allele (55%) in both subpopulations studied and for the OPC the genotype MM and the M allele presented higher frequency (Table 1).
This is consistent with the frequencies found by Santos et al [12] in three of the four breeds studied in Brazil; among them Pantaneira (MM: 58%, MN: 23%, NN: 19%, M: 70%), Suffolk (MM: 80%, MN: 16%, NN: 4%, M: 88%) and the Ile de France (MM: 82%, MN: 12%, NN: 6%, M: 88%) but not with what was found in the Bergamacia breed (MM: 28%, MN: 34%, NN: 38%, M: 45%). In Afshari Iranian sheep the M and N alleles are reported with frequencies of 74% and 26%, respectively [16] and in the genotypic frequencies found, the homozygote NN presented a higher frequency (10%) than the one shown here; the others genotypes had frequencies of 57 and 33% for MM and NN, respectively.
On the other hand, in the Zel breed, the allelic frequencies were 85.5% for M and 14.5% for N [17], frequencies for genotypes MM (75%) and NN (4%) lower than that found in the OPC. In autochthonous sheep of Pakistan Balkhi, Kajli and Beetal the frequencies of the M allele were 88, 86 and 100%, respectively, with the most frequent MM genotype (Balkhi: 76%, Kajli: 74%, Beetal: 100%) and absence of the NN genotype in the Balkhi and Beetal breeds and with low frequency (2%) in Kajli [18]. The frequency of the N allele in the Kurdi race was higher (12%) than that found in the OPC with the absence of the NN genotype and higher MM frequency (76%) [19]. It is noteworthy that the M allele of this locus has been associated with high weights at birth [15].
For the CAST locus, the PCR-RFLP and PCR-SSCP tech-niques were compared as alternatives for genotyping. It was determined that the A and C alleles obtained by SSCP could be digested with the MspI enzyme, also, that the AA, AC and CC were similar to the MM genotype determined by RFLP, similarly the MN genotype is similar with the AB and BC genotypes (by SSCP) and finally, the NN genotype agrees with the BB genotype [11,17]. In another locus of the CAST gene (exons 10, 11 and intron 10) in 80 animals of the OPC breed, the allelic frequencies found were 93.7% for A and 6.3% for B, with genotype frequencies of 88.75, 10 and 1.25% for AA, AB and BB respectively [20].
The value of He at the CAST locus was lower than that found in the Dalagh [11] (49%) and Zel [16] (25%), Pantaneira (42%), Suffolk (21%) and Ile de France (21%) and Bergamacia (50%) [12] Balkhi (21%) and Kajli (24%) [18] breeds, but higher than that reported in OPC (11%) [20] and the Kurdi sheep (7.9%) (19). Similarly, in the present investigation a non-significant excess of heterozygotes was found, as was EHW. This indicates that no selection has been made in favor of any of the alleles of this locus, or the absence of genetic improvement programs assisted by molecular tools. On the other hand, Molano [20] in the same racial group studied here, presents a non-significant heterozygous deficit and absence of the theoretical proportions of the EHW. Contrary results are presented in Dalagh sheep [11] and Zel [17] the Pantaneira, Ile de France and Bergamacia [12] where the authors found heterozygous deficit with significant deviations of EHW (Table 2).
For the CAPN locus (AF309634.1) a variation located between exons 5 and 6 was studied, this consisted of a synonymous transition of T/C in nucleotide 44 of the gene [11,17]. Variations in this locus have been associated with growth characteristics and channel quality in sheep [21,22]. Contrary to what has been reported here, three breeds of sheep from Egypt (Barki, Rahmani, and Ossimi) present the CC genotype and the C allele as the most frequent and with the absence of the TT genotype in the Barki and Ossimi breeds [22] (Table 1). In another locus (CAPN316) of the CAPN gene but in the OPC breed, the allelic frequencies found were similar (A: 54.4% and B: 45.6%) with greater frequency of the heterozygous genotype (38.75%) than of the homozygotes (AA: 35% and BB: 26.25%) [20].
In Merino sheep, three alleles are reported for this gene, with the A1 allele and A1B1 genotype being the most frequent, with values of 46.7 and 41.9%, respectively [23]. In Baluchi sheep, three band patterns are reported with genotypic frequencies of 8.2 (G1), 89 (G2) and 3% (G3) (11). In Bandur sheep, they report two alleles with frequencies 82 (A) and 18% (B), and two genotypes AA (67%) and AB (33%) [24], similarly, only the AA genotypes were found in Zel sheep. (69%) and AB (31%) with allelic frequencies of 84.5 and 15.5 for A and B, respectively [ ] and in Kurdi sheep [19] the allele frequencies for A and B were 96 and 4% with the absence of the genotype BB and greater frequency of the AA genotype.
The research cited and the present work used the PCR-SSCP genotyping methodology for the CAPN locus, despite the simplicity of the technique, in several reports, the mobility patterns of the bands were different. Some reasons that could explain why more than two bands can be observed in the single-stranded DNA strands being the same sequence, are related to the molecular stability of the chain that can be affected by an excess of primers, a slow handling after the denaturation that cause a bio-molecular pairing or re-association [25]; and by electro-phoresis conditions, where factors such as temperature, run time, buffer concentration, gel composition, position of the SNP in the analyzed fragment as well as fragment size, can influence the sensitivity of the technique [26].
The expected heterozygosity found in this study for the CAPN locus is higher than that reported in the breeds Bandur (29.5%) [24], Zel (26%) [17] and Kurdi (7%) [19], but lower than that reported by Molano [20] in OPC (49%). On the other hand, a non-significant excess of intra-population heterozygotes was found at this locus, but when analyzing the entire study population (Table 2), this excess was significant with deviations from the EHW (p<0.05). This could be attributed to the lack of inbreeding due to the existence of gene flow, as well as the rapid crossing of migrating females and males with the individuals residing in the subpopulations. In contrast, heterozygous deficits without deviations of EHW occur in OPC [20], and in the Zel breed, this deficit caused deviations of EHW (p<0.05) (17). Likewise, in the Kurdi and Bandur breeds, the excess of heterozygotes found did not cause significant deviations of EHW [19,24].
The analysis of molecular variance showed that the variation found is only due to differences within individuals, which is confirmed by the lowest value of FST found (0.002, p=0.653), which indicates a low population structure with gene flow among the subpopulations studied [5], in addition, the values of negative and not significant FIS and FIT (p> 0.05) contrast with what was reported in OPC (FIS=0.183) [20], which could be explained by the different origin of the animals evaluated as well as differences in the management of production systems with respect to their systems or crossing schemes.
Finally, the results of the present study showed that the loci studied have good variability that is necessary to conserve. Additionally, the allelic frequencies found are considerably high, so these results could be used in future gene-assisted selection plans, in order to increase the productivity of the sheep systems that use the OPC as a genetic base.











 texto em
texto em 


