Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista MVZ Córdoba
Print version ISSN 0122-0268On-line version ISSN 1909-0544
Rev.MVZ Cordoba vol.24 no.2 Córdoba May/Aug. 2019
https://doi.org/10.21897/rmvz.1642
Research article
Selection markers in Pampa Rocha piqs: a comparison with autochthonous breeds of Spain and Portugal
1Universidad de Zaraqoza. Facultad de Veterinaria, Departamento de Producción Animal y Ciencia de los Alimentos. Miguel Servet 177, C.P. 50013, Zaragoza, España.
2Universidad de la República, Facultad de Veterinaria, Departamento de Genética y Mejora Animal. Lasplaces 1550. C.P. 11600, Montevideo, Uruquay.
3Universidad de Zaragoza. Facultad de Veterinaria, Laboratorio de Citoqenética y Genética Molecular. Miquel Servet 177, C.P. 50013, Zaragoza, España.
Objective.
The analysis of selection markers allows to obtain information about the evolutive story of a particular breed or line and allows also to evaluate the usefulness of those markers for breeding programs. We have analyzed SNPs in four genes of the creole pig breed Pampa Rocha and we have compared their allelic frequencies with the allelic frequencies of diverse autochthonous breeds of Spain and Portugal and also with Piétrain pigs and wild boars.
Results.
The results of the analysis show that Pampa Rocha pigs have similar allelic frequencies with the autochthonous breeds of Spain and Portugal especially in the case of IGF2 and also, but not so coincident, in the case of PEPCK-C. However, they differ considerably for MC4R, and also, but in a lower extent, for PRKAG3. We discuss in this work the usefulness of our results for breeding of Pampa Rocha pigs.
Conclusions.
Our results demonstrate the peculiarity of the Pampa Rocha breed regarding the markers studied.
Keywords: IGF2; MC4R; PRKAG3; PEPCK; Sus scrofa (Source: CAB; BioThesaurus)
Objetivo.
El análisis de marcadores de selección permite obtener datos de la vida evolutiva de una raza o línea y permite también evaluar la conveniencia o no de su uso en programas de mejora genética. Hemos evaluado SNPs en cuatro genes (IGF2, MC4R, PRKAG3 y PEPCK-C), que tienen importantes efectos fenotípicos, en cerdos de la raza Pampa Rocha, una raza criolla, y hemos comparado sus frecuencias alélicas con cerdos de diversas razas autóctonas y líneas de España y Portugal no sometidas a selección así como con jabalíes y cerdos de la raza Piétrain.
Resultados.
Los resultados de los análisis muestran una similitud de frecuencias alélicas entre los cerdos de la raza Pampa Rocha y los cerdos autóctonos de la península ibérica sobre todo en el gen IGF2 y, en menor medida en el gen PEPCK-C. Sin embargo difieren considerablemente en el caso del marcador MC4R y, también en menor medida, en PRKAG3. En el trabajo se discute el uso potencial de los resultados obtenidos para orientar la selección genética de cerdos de la raza Pampa Rocha.
Conclusiones.
Nuestros resultados demuestran la peculiaridad de la raza Pampa Rocha con respecto a los marcadores estudiados.
Palabras clave: IGF2; MC4R; PRKAG3; PEPCK; Sus scrofa (Fuente: CAB; BioThesaurus)
INTRODUCTION
Pig selection has been oriented in the last decades toward the obtaining of lean animals. The success of this process can be exemplified by the reduction in conversion indexes from about 4.3 and above in the sixties down in the present time to less than 2.2 in the most efficient breeds such as the Piétrain one 1. In this process, backfat thickness (BT) has been used as selection criteria to breed animals. The reduction in BT has been spectacular. Nowadays, BT is considered to be at the lowest physiological limit in very selected breeds. Reduction in BT has been accompanied by a reduction in intramuscular fat (IMF). As a consequence of reduction in IMF, some very important meat quality traits such as juiciness, taste or water holding capacity, have been very negatively affected 2,3. Present pigs are very efficient biological machines converting feed into meat but they render meat with low sensorial quality.
There exists another market strategy in some European industrialized countries to counteract this trend: the use of autochthonous breeds not submitted to selection toward leanness to obtain high quality meat derived products directed to highly priced markets 4. The paradigm of the success of this strategy is the Iberian breed, whose cured derived products achieve very high prices (up to u$ 788 for just one whole cured ham in the most prestigious brands).
Complex traits, such as IMF, are the consequence of the effects of many minor genes. However, there exist also some genes whose effects are especially high in the shaping of this trait. These genes, known as major genes, can be used to accelerate the shaping of desired phenotypes. There exist a few known major genes involved in determining of fat content (both IMF and BT): RYR1, the halothane gene, IGF2, insulin like growth factor 2 5, MC4R, melanocortin receptor 4 6, or PRKAG3, a regulatory subunit of AMP-activated protein kinase 7 are some of the best known.
Recently, a SNP was discovered in the PEPCK-C, cytosolic phosphoenolpyruvate carboxykinase, that has opposite and remarkable effects on BT and IMF 1. These markers can be very useful for various purposes: on one side to investigate the selection history of a breed or to assign individuals to a particular breed and, on the other side, to introgress them (in the case they are already present) in not selected breeds to improve, mostly, productive traits.
Pampa Rocha is a creole pig from Uruguay whose genetic origin are the pigs introduced by Spanish and Portuguese colonists. These pigs had been already crossed with Asian populations introduced several centuries ago to improve mostly maternal traits such as prolificacy. Later on, Pampa Rocha pigs have been crossed with Berkshire and Poland China breeds 8. Pampa Rocha females have very interesting maternal traits and grazing adaptability. Their crossbreeding with Duroc males gives an offspring with very improved growth and weight gain properties. Duroc males are also crossed with Iberian sows to improve productive traits 9. The cured products derived from pigs of this cross are among the best Iberian products, provided that the Duroc line used is not lean.
The objective of the present work was to investigate the presence of alleles associated with selection in Pampa Rocha pigs. The mutations IGF2 (5), MC4R (6), PRKAG3 (7) and PEPCK (1) were analyzed by various methods and were also studied in wild boars, Piétrain pigs and some breeds or lines of native pigs from Spain and Portugal. Finally, the usefulness of this analysis was evaluated to improve the commercial value of the pigs of the Pampa Rocha breed.
MATERIALS AND METHODS
Animal material. Frozen muscles samples from wild boars were supplied by several hunting friends of our Department. Frozen muscle samples from Iberian pigs were obtained from COVAP (Pozoblanco, Spain) and CENSYRA (Badajoz, Spain). Dr. Carballo García (Universidad de Vigo, Spain) supplied us frozen muscle samples from Porco Celta and hair samples from Bízaro pigs. Hair samples from Pampa Rocha pigs were obtained from animals belonged to the breeding group of the Pig Production Unit of the Southern Regional Center of the Experimental Station of the Faculty of Agronomy (University of the Republic) located in Progreso, Canelones, Uruguay. Semen from Piétrain pigs was obtained from Agropecuaria OBANOS S.A. (Marcilla, Spain).
DNA extraction. DNA was extracted from muscle, semen or hair follicles using the RealPure Genomic DNA Extraction kit from Durviz (Valencia, Spain) or the GenomicPrepTM DNA isolation kit from Amersham Biosciences (Little Chalfont, UK), according to the manufacturer's instructions. DNA concentration in the extract was determined using a NanoDrop 1000 spectrophotometer (Thermo Fischer Scientific, Wilmington, DE, USA).
Genotyping. Genotyping of IGF2, MC4R, PRKAG3 and PEPCK-C was performed by Real Time PCR as described in Burgos et al 10, Burgos et al 11, Galve et al 12 and Latorre et al 1 respectively.
RESULTS
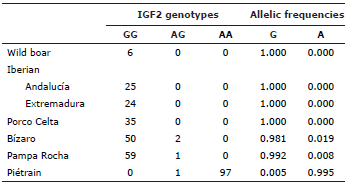
Table 1 presents the analysis of the IGF2 genotype of the Pampa Rocha pigs and of the autochthonous breeds of Spain and Portugal. The allele G is the wild type one. The allele A represents in the Piétrain population over 99% of total alleles. Pampa Rocha pigs are, on the other side, almost all homozygous GG. Only one animal is heterozygous. In this sense, the wild boars, the Spanish breeds and lines and the Portuguese Bízaro pigs are very similar to the Pampa Rocha ones: Porco Celta pigs and pigs of the two Iberian lines are all homozygous GG whereas only two Bízaro pigs are heterozygous being the rest also homozygous GG.
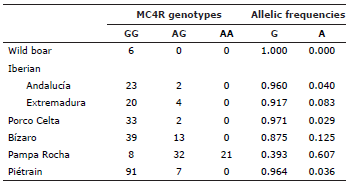
The results of the genotyping of MC4R is shown if table 2. It is no surprise, that the A allele is almost fixed in Piétrain pigs, a breed characterized by a strong contention eating feed 13. It is curious that wild boars present also this genotype. The same occurs with all Spanish and Portuguese breeds, although the presence of A alleles is in all these cases far from negligible. The MC4R genotypes of Pampa Rocha pigs differ considerably from all these breeds and lines because the A alleles are the most abundant ones.
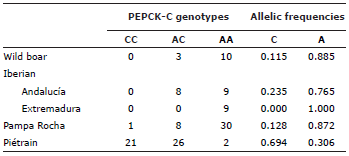
Finally, the PEPCK-C genotypes are shown in table 4. In this table data from Bízaro and Porco Celta pigs are not included. Unfortunately, when we discovered this SNP (1) we had no DNA from Bízaro and Porco Celta pigs available. All breeds and lines not selected for lean content, included the Pampa Rocha one, have preponderantly the A allele. On the opposite side, the Piétrain pigs have more C alleles.
DISCUSSION
The allele A of IGF2 was originated in Asian pigs and is responsible of 10-20% of the phenotypic variability in fat traits 5. Therefore, it is an SNP of extraordinary relevance due to these large phenotypic effects in relevant productive and morphological traits 10,14. It is one of the few markers that can be used itself as selection criteria. Its use in Pampa Rocha is limited by the almost absence of A alleles. A alleles are associated simultaneously to lower levels of backfat thickness and larger IMF content. If a breeding scheme consisting in crossing Pampa Rocha sows (mostly GG) with AA Duroc males would be implemented, the present IGF2 status of Pampa Rocha pigs would be very adequate. This occur because the imprinted nature of the IGF2 locus 15; only the alleles received from the male (A alleles) would be expressed in the offspring, making the maternal allele irrelevant for the F1. However, this G allele is very relevant for maternal traits; indeed, GG genotype in the sows result in 0.5-1 piglet more in each parity 16. This is due to the large fattening effect of the G allele and the well-known relation between adequate fattening of sows, larger milk production capacity and increased number of live piglets in the offspring.
MC4R encodes a receptor involved in the neural circuits that regulate food intake 17,18 The A allele is associated to increased food intake and, through this, to increased BT and enhanced growth rate 6,19,20. Using this SNP as selection criteria could direct Pampa Rocha pigs to enhanced fattening (A alleles) or to more leanness (G alleles). As the fattening of Pampa Rocha pigs is already high enough, we think that it could be advisable to select animals of the GG genotype to improve productive traits of this breed.
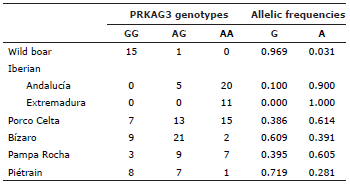
The mechanism by which the SNP in the PRKAG3 gene, described by Ciobanu et al 7, has effects on fat related traits is unknown. PRKAG3 encodes a regulatory subunit of AMP activated protein kinase, which is involved in the determination of glycogen content in striated muscle and, through this, of ultimate pH postmortem in meat. The A allele is associated to enhanced BT values 12,21. The selection of A or G alleles is a complex decision because it affects several phenotypic traits in a complex manner. However, the near equilibrated presence of G and A alleles of this SNP makes its selection in Pampa Rocha pigs a straightforward process.
Finally, the A allele of PEPCK-C is associated to larger IMF content, lower BT, and less exudation 1, an uncommon phenotype very valuable for the production of quality meat. This means that meat from Pampa Rocha pigs could still be improved if the few animals that contain C alleles would be excluded from reproduction.
The scarcity of analyzed samples did not allow us to study haplotypes combining the four genes. A very important major gene, RYR1, has not been considered in our study because their involvement in the apparition of PSE meat 22, although it is very improbable to find variation in this marker because it appeared originally in the Piétrain breed. Pampa Rocha pigs have common allelic frequencies with Spanish and Portuguese autochthonous breeds for some genes associated to breeding processes. This is especially so in the case of IGF2, whose G allele is fixed in all breeds or lines not submitted to artificial selection, including the Pampa Rocha one, whereas the A allele is almost fixed in Piétrain pigs, the paradigm of muscularity.
The allelic frequencies for the PEPCK-C marker of all not selected breeds show a similar pattern although there is some variability in this marker in all tested breeds. Pampa Rocha is close to breeds of Celtic origin, and especially to Porco Celta, in the allelic frequencies of PRKAG3. On the contrary, Pampa Rocha differs from all autochthonous breeds studied in this work in the allelic frequencies of the MC4R marker, showing the uniqueness of this breed probably due to its different evolutive story.
Acknowledgments
This work was supported by project AGL2015-66177-R to Pascual López-Buesa. Silvia LLambí was partially financed by CSIC-UdelaR.
REFERENCES
1. Latorre P, Burgos C, Hidalgo J, Varona L, Carrodeguas JA, López-Buesa, P. c.A2456C-substitution in Pck1 changes the enzyme kinetic and functional properties modifying fat distribution in pigs. Sci Rep. 2016; 6:19617. https://doi.org/10.1038/srep19617 [ Links ]
2. Font-i-Furnols M, Tous N, Esteve-Garcia E, Gispert M. Do all the consumers accept the marbling in the same way? The relation between visual and sensory acceptability of pork. Meat Sci. 2012; 91(4):448-453. https://doi.org/10.1016/j.meatsci.2012.02.030 [ Links ]
3. Reina R, López-Buesa P, Sánchez del Pulgar J, Ventanas J, García C. Effect of IGF-II (insulin like growth factor-II) genotype on the quality of dry-cured hams and shoulders. Meat Sci. 2012; 92(4):562-568. https://doi.org/10.1016/j.meatsci.2012.05.027 [ Links ]
4. Pugliese C, Sirtori F. Quality of meat and meat products produced from southern European pig breeds. Meat Sci. 2012; 90(3):511-518. https://doi.org/10.1016/j.meatsci.2011.09.019 [ Links ]
5. Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collete C, Moreau L et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003; 425(6960):832-836. https://doi.org/10.1038/nature02064 [ Links ]
6. Kim KS, Larsen N, Short T, Plastow G, Rothschild MF. A missense variant of the melanocortin-4-receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mam Genome. 2000; 11(2):131-135. https://doi.org/10.1007/s003350010025 [ Links ]
7. Ciobanu D, Bastiaansen J, Massoud M, Helm J, Woollard J, Plastow G, Rothschild M. Evidence for new alleles in the protein kinase adenosine monophosphate-activated g3-subunit gene associated with low glycogen content in pig skeletal muscle and improved meat quality. Genetics. 2001; 159(3):1151-1162. PMID:11729159 [ Links ]
8. Montenegro M, Llambí S, Castro G, Barlocco N, Vadell A, Landi V et al. Genetic characterization of Uruguayan Pampa Rocha pigs with microsatellite markers. Genet Mol Biol. 2015; 38(1):48-54. https://doi.org/10.1590/s1415-475738120140146 [ Links ]
9. Fuentes V, Ventanas S, Ventanas J, Estévez M. The genetic background affects composition, oxidative stability and quality traits of Iberian dry-cured hams: Purebred Iberian versus reciprocal Iberian x Duroc crossbred pigs. Meat Sci. 2014; 96(2):737-743. https://doi.org/10.1016/j.meatsci.2013.10.010 [ Links ]
10. Burgos C , Galve A, Moreno C, Altarriba J, Reina R, García C, López-Buesa. The effects of two alleles of IGF2 on fat content in pig carcasses and pork. Meat Sci. 2012; 90(2):309-313. https://doi.org/10.1016/j.meatsci.2011.07.016 [ Links ]
11. Burgos C, Carrodeguas JA, Moreno C, Altarriba J, Tarrafeta L, Barcelona JA et al. Allelic incidence in several pig breeds of a missense variant of pig melanocortin-4 receptor (MC4R) gene associated with carcass and productive traits; its relation to IGF2 genotype. Meat Sci. 2006; 73(1):144-150. https://doi.org/10.1016/j.meatsci.2005.11.007 [ Links ]
12. Galve A, Burgos C, Varona L, Carrodeguas JA, López-Buesa P. Allelic frequencies of PRKAG3 in several pig breeds and its technological consequences on a Duroc 9 Landrace-Large White cross. J Anim Breed Genet. 2013; 130(5):382-393. https://doi.org/10.1111/jbg.12042 [ Links ]
13. Saintilan R, Mérour I, Schwob S, Sellier P, Bidanel J, Gilbert H. Genetic parameters and halothane genotype effect for residual feed intake in Piétrain growing pigs. Livest Sci. 2011; 142(1-3):203-209. https://doi.org/10.1016/j.livsci.2011.07.013 24(1) Mayo - Agosto 2019 [ Links ]
14. Van den Maagdenberg K, Stinckens A, Claeys E, Buys N, De Smet S. Effect of the insulin-like growth factor-II and RYR1 genotype in pigs on carcass and meat quality traits. Meat Sci. 2008; 80(2):293-303. https://doi.org/10.1016/j.meatsci.2007.12.008 [ Links ]
15. Li C, Bin Y, Curchoe C, Yang L, Feng D, Jiang Q et al. Genetic imprinting of H19 and IGF2 in domestic pigs (Sus scrofa). Anim Biotechnol. 2008; 19(1):22-27. https://doi.org/10.1080/10495390701758563 [ Links ]
16. Stinckens A, Mathur P, Janssens S, Bruggeman V, Onagbesan OM, Schroyen M et al. Indirect effect of IGF2 intron3 g.3072G>A mutation on prolificacy in sows. Anim Genet. 2010; 41(5):493-498. https://doi.org/10.1111/j.1365-2052.2010.02040.x [ Links ]
17. Krashes MJ, Lowell BB, Garfield AS. Melanocortin-4 receptor-regulated energy homeostasis. Nat Neurosci. 2016; 19(2):206-219. https://doi.org/10.1038/nn.4202 [ Links ]
18. Shen WJ, Yao T, Kong X, Williams KW, Liu T. Melanocortin neurons: Multiple routes to regulation of metabolism. Biochim Biophys Acta. 2017; 1863(10 Pt A):2477-2485. https://doi.org/10.1016/j.bbadis.2017.05.007 [ Links ]
19. Davoli R, Braglia S, Valastro V, Annaratone C, Comella M, Zambonelli et al. Analysis of MC4R polymorphism in Italian Large White and Italian Duroc pigs: association with carcass traits. Meat Sci. 2012; 90(4):887-892. https://doi.org/10.1016/j.meatsci.2011.11.025 [ Links ]
20. Switonski M, Mankowska M, Salamon S. Family of melanocortin receptor (MCR) genes in mammals-mutations, polymorphisms and phenotypic effects. J Appl Genet. 2013; 54(4):461-472. https://doi.org/10.1007/s13353-013-0163-z [ Links ]
21. Skrlep M, Kavar T, Santé-Lhoutellier V, Candek-Potokar M. Effect of I199V polymorphism at PRKAG3 gene on carcass and meat quality traits in Slovenian commercial pigs. J Muscle Foods. 2009; 20(3):367-376. https://doi.org/10.1111/j.1745-4573.2009.00158.x [ Links ]
22. Salas RC, Mingala C. Genetic Factors Affecting Pork Quality: Halothane and Rendement Napole Genes. Anim Biotechnol. 2017; 28(2):148-155. https://doi.org/10.1080/10495398.2016.1243550 [ Links ]
How to cite (Vancouver) Burgos SC, Llambí DS, Hidalgo GJ, Montenegro SM; Arruga LV, López-Buesa P. Selection markers in Pampa Rocha pigs: a comparison with autochthonous breeds of Spain and Portugal. Rev MVZ Cordoba. 2019; 24(2):7198-7202. DOI: https://doi.org/10.21897/rmvz.1642
Creative Commons Attribution 4.0 International License This article is distributed under the terms of the (https://creativecommons.org/licenses/by-sa/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source. 
Received: October 01, 2018; Accepted: March 01, 2019; Published: April 01, 2019











 text in
text in