Introduction
Cape gooseberry (Physalis peruviana, L.) is an exotic fruit that belongs to the Solanaceae family. It is native to the Andean region and its fruits are recognized by their anti-inflammatory, antioxidant and anticancer properties commonly used to manage diabetes and hypertension [1-3]. Colombia is the world's top producer and exporter with annual sales of $ 30 million dollars for 2017 [4, 5]. However, cape gooseberry production is limited due to problems in fruit quality [6] and losses due to the vascular wilt disease [7], caused by the fungus Fusarium oxysporum f. sp. physali (Foph) [8]. In order to understand the molecular mechanisms of important agronomic traits, recent genome wide association studies have been conducted by Osorio-Guarín etal. 2016 [9] and Garcia-Arias etal. 2018 [10] in which 27 and 34 candidate genes were associated to resistance response and fruit quality, respectively.
Despite the high comercial value of cape gooseberry, scarce information regarding gene function is available. Gene function studies are essential to better understand the molecular mechanisms that regulate particular traits generating molecular markers that can assist breeding programs. Consequently, the implementation of techniques for identifying gene functions in P. peruviana is essential.
Techniques such as stable transgene overexpression, DNA transfer, transposon-based mutagenesis and stable RNA interference (RNAi) have been used to analyze the functions of candidate genes [11]. However, these techniques have some disadavantages such as being relatively expensive, time-consuming and laborious [12]. Virus-induced gene silencing (VIGS) is a rapid tool for functional analyses used in different crops and non-model species such as wheat [13], tomato [14] and rye [15]. In addition, this method allows silencing different genes simultaneously in different plant organs like roots and leaves [16-17].
The VIGS technique employs a recombinant virus to specifically reduce endogenous gene activity through a plant defense mechanism called post-transcriptional gene silencing (PTGS) [18]. The binary viral vector used in VIGS contains its viral genome and a fragment of the target gene of the host plant; then the recombinant vector is transferred into the plant via Agrobacterium tumefaciens. Once the virus has spread in the plant, it produces a double-stranded RNAs (dsRNAs) that contains the target gene to be silenced [19]. Dicer-like proteins cleave these viral dsRNAs into short interfering RNAs (siRNA) [20, 21]. The siRNA integrates into the RNA-induced silencing complex (RISC) that guides the cleavage of the complementary viral RNA, causing the transiently loss of the gene target function [22].
A commonly used virus in VIGS is the tobacco rattle virus (TRV) [23, 24]. This virus has two genomes: 1) pTRV1, which is essential for the viral movement that causes the infection, and 2) pTRV2, which has genes encoding coat and nonstructural proteins; these are not essential for plant infection and are replaced with multiple cloning sites for inserting fragments of the target genes to be silenced [23, 25].
The aim of this study is to optimize the conditions for a TRV-based VIGS protocol and to provide the first gene function screening of Physalis peruviana using the phytoene desaturase (PDS) gene, in which down-regulation shows partial photo-bleaching in green tissues [26].
Materials and methods
Plant material
Cape gooseberry (accession 12U134) seeds were obtained from the germplasm collection of Corporación Colombiana de Investigación Agropecuaría (Agrosavia). Forty plants were grown in individual pots with fertilized soil in a greenhouse at 25 ◦ C under a photoperiodic lighting of 16 h light/8 h dark.
Primer design
The coding sequence (CDS) sequence of the PDS gene from tomato (Solanum lycopersicum) was acquired from the National Center for Biotechnology Information NCBI (NM_001247166.2). A BLASTn program was used to compare this gene against the cape gooseberry root/stem transcriptome (NCBI Bioproject ID No. PRJNA67621). The transcript TR18226 aligned with other Solanaceae PDS homologs resulted in the identification of the PpPDS (Physalis peruviana - Phytoene desaturase) sequence; and the best fragment for silencing inside the TR18226 transcript was selected using the VIGS-SGN tool [27].
The primers used to amplify the fragment of the PpPDS gene were designed with the software PRIMER3 v.0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) based on a variety of parameters such as primer size of 22 base pairs (bp), primer Tm of 60 ◦ C, primer GC % of 45. Finally, we added adaptors at the 5'-end of the specific primers to create complementary overlaps useful to anneal the vector and insert [28]. The sequences are: 5'-adaptor: CGACGACAAGACCCT - primer: TGCTTTTGTGTTTGCCACTC-3' and 5'-adaptor: GAGGAGAAGAGCCCT - primer: GGTTCACAACCTGGCACAGT-3'.
PDS amplification
RNA from young leaves was isolated using the RNeasy plant Mini Kit (Qiagen, Hilden, Germany). The RNA was treated with the TURBO DNase (Ambion) to remove the genomic DNA and transcribed into cDNA using the iScript cDNA Synthesis kit (BioRad) following the manufacturer's instructions. The PpPDS gene was amplified using the cDNA as template with the following parameters: initial denaturation at 95 ◦ C for 5 min, 35 cycles of 94 ◦ C for 30 s, 57 ◦ C for 30 s and 72 ◦ C for 30 s, followed by a final extension at 72 ◦ C for 5 min. Finally, the PCR product was purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer's instructions.
Plasmid construction
Cells containing the pTRV2 (pYL156) and the pTRV1 (pYL192) vectors were grown in liquid LB medium at 37 ◦ C overnight at 100 rpm, and then plasmid DNA was extracted using the PureLink HQ Mini plasmid Purification Kit.
The purified PpPDS gene was inserted into the pTRV2 (pYL156) vector from tobacco rattle virus (TRV), previously described by Liu et al. 2002 [29], using the ligation independent cloning (LIC) procedure. In this procedure, the T4 DNA polymerase with exonuclease activity 3' 5' removed one of the complementary strands of the adaptor in the vector and in the insert to create overlapping sticky ends.
Five hundred nanograms of the pTRV2 vector (pYL156) were linearized with the PstI enzyme at 37 ◦C for 1 h. The linearized product was treated with T4 DNA polymerase (Invitrogen) in 1x reaction buffer containing 5 mM dTTP at 22 ◦ C for 30 min, followed by 20 min of inactivation at 70 ◦ C. In parallel, the same treatment was carried out for the PpPDS product but with 5 mM dATP. Both products were mixed at a 1:1 ratio and incubated for 2 min at 65 ◦C, and later for 10 min at room temperature to allow the annealing of the complementary sticky ends; the final result was the pTRV2:PpPDS vector.
A pTRV2: NobPDS vector with the PDS gene of the species Nicotiana obtusifolia (desert tobacco) constructed by Natalia Pabón-Mora from the EvoDevo research group of the Universidad de Antioquia was used as a positive control. To amplify the Nicotiana PDS, the following primers were used: 5'-CGACGACAAGACCCT-CACCTGGGAGTTCCTGTGATAAAT-3' and 5'-GAGGAGAAGAGCCCT-GTGTACAACGCTAATTCAGCG-3'.
Cloning and transformation
In this phase, six μL of the constructed vector were transformed into 20 μL of chemically competent Escherichia coli TOP10 cells. Cells were resuspended in 250 μL of S.O.C medium and incubated at 37 ◦C for 1 h. Subsequently, 50 μL of the transformation mixture was plated onto a LB agar plate supplemented with kanamycin (50 μg/mL), and incubated at 37 ◦C overnight. Transformants were tested by PCR amplification using PpPDS primers. The PCR product from the E. Coli positive transformants were submitted to Sanger sequencing to verify the insert sequence before proceeding with the experiment.
Further, 2 μL of the construct was transformed into 50 μL of home-made electro-competent Agrobacterium tumefaciens strain GV3101 cells by electroporation using the following program: 2.1 kV, 100 Ω, and 25 pF in a MicroPulser (Bio-Rad) electroporator using 0.1 cm gap electroporation cuvettes. Cells were resuspended in 950 μL of S.O.C medium and were then incubated at 28 ◦C for 1 h. An aliquot of 70 μL of transformation mixture was plated on LB agar supplemented with Kan50, Gent25 and Rif25. The resulting Agrobacterium transformants were grown in liquid LB medium at 28 ◦C during 48 h at 200 rpm; and the plasmid DNA was extracted using the PureLink HQ Mini plasmid Purification Kit and checked through PCR amplification employing the pTRV2 and PpPDS primers and the products were sequenced by Sanger sequencing. The pTRV1 (pYL192) and pTRV2:NobPDS were transformed in Agrobacterium using the same procedure.
Preparation of virus inoculum and inoculation of cape gooseberry plants
Agrobacterium colonies with the pTRV1, pTRV2:PpPDS and pTRV2:NobPDS vectors were grown separately in 5 mL of liquid LB medium with Kan50, Gent25, and Rif25, overnight at 28 ◦ C at 200 rpm in darkness. The cultures were then transferred to 50 mL of liquid LB medium with Kan50, Gent25, Rif25, 1 M MES, and 0.1 M acetosyringone, and shaken overnight at 200 rpm at 28 ◦C in darkness until they reached an optical density (OD600) of 1.5. Cells were harvested at 4 ◦C at 4 000 rpm and resuspended in infiltration medium (1 M MgCl2, 1 M MES, and 0.1 mM acetosyringone) into two different concentrations (OD600 of 1.0 and 1.5). Then pTRV1 and pTRV2 were mixed in a 1:1 ratio. The mixtures produced were: 1) pTRV1 and pTRV2:PpPDS at 1.0 OD, 2) pTRV1 and pTRV2:PpPDS at 1.5 OD, and 3) pTRV1 and pTRV2:NoPDS at 1.0 OD. Finally, the mixtures were incubated at room temperature for 3 h before the infiltration process.
Two treatments, two negative controls and one positive control were used during the agroinfiltration step: 1) Five plants were treated with infiltration solution (negative control), 2) Five plants were infiltrated with pTRV1 (negative control), 3) Ten plants were infiltrated with pTRV1-TRV2:PpPDS at OD: 1.0, 4) Ten plants were infiltrated with pTRV1-pTRV2:PpPDS at OD: 1.5, and 5) Ten plants were infiltrated with pTRV1-pTRV2:NobPDS (positive control) at OD: 1.0. Three leaves per plant were infiltrated with a 1 mL syringe without the needle and then placed in a greenhouse under the following conditions: 16 h light/8 h dark cycle, 60 % of relative humidity and a temperature of 25 ± 2 ◦ C. The experiment was monitored during 47 days post-inoculation (dpi).
Plants with photo-bleaching symptoms were counted during the first 21 dpi. The PpPDS gene-silencing efficiency was calculated as the percentage of plants showing a photo-bleached phenotype in at least one leaf out of the total number of plants tested. In addition, we counted the bleached leaves over the inoculated leaves.
RNA isolation and expression analysis
Small pieces of leaves were frozen in liquid nitrogen and stored at -80 ◦ C. Total RNA was isolated from mutated and non-mutated leaves from each treatment (OD:1.0, OD:1.5, pTRV1 and infiltration solution) using the RNAeasy plant Mini Kit (QIAGEN, Germany) according to the manufacturer's instructions, and the final elute was adjusted to 30 μL. NanoDrop 1 000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, U.S.A), was used to quantify the total RNA-DNA. To remove genomic DNA, 10 μg of the previous quantification was treated with TURBO DNase (Ambion). For the cDNA synthesis, 500 ng of RNA was used as a template.
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) was used to corroborate if the silencing of the PpPDS gene was successful. New primers were designed to prime outside the homology region between the VIGS vector and the previous inserted PDS region. The sequences of the primers were: 5'-TTTACGCAAGTGGCCAAAC-3' and 5'-CCAGGGCAAAAGCAAATAAA-3'. The conditions for the RT-PCR were: initial denaturation at 95 ◦ C for 5 min, 35 cycles of 94 ◦ C for 30 s, 57 ◦ C for 30 s and 72 ◦ C for 30 s, followed by a final extension at 72 ◦ C for 5 min. The reaction was checked through electrophoresis on a 1 % agarose gel.
In addition, real-time quantitative PCR using the QTM SYBR® Green supermix (BioRad) reagent was conducted in an iQ5 Thermocycler (Bio-Rad) using the PpPDS primers excluding the adaptor sequence at the following conditions: an initial denaturation step of 95 ◦ C for 3 min, then 40 cycles of 95 ◦ C for 15 s followed by 56 ◦ C for 1 min. Melting curves were generated to check the amplification specificity. The qPCR reaction was carried out using two plants for each of the pTRV1-TRV2:PpPDS treatments. The gene a-tubulin was used as endogenous control.
Results and discussion
Plant genetic transformation has been used as a robust tool for the genetic engineering of many several crops such as tobacco, tomato, soybean, rice, and wheat. However, transformation protocols in non model species such as P. peruviana are less available. In order to validate the function of genes in this species, this study implemented VIGS as a tool for silencing genes in cape gooseberry.
Plasmid construction
To standardize the VIGS protocol, a PDS gene was used as a target gene because when it is silenced a visible plant phenotype is produced. The non-expression of the PDS gene causes a loss of carotenoids, and as a consequence, plants show the photo-bleached phenotype that has been reported in tomato and tobacco [30]. Liu & Page (2008) [31] reported that the insert length to silence effectively PDS in Nicotiana benthamiana ranges between 200 bp and 1 300 bp and the selected gene region should exclude 3' and 5' ends of the PDS gene to be more efficient. The cape gooseberry reference genome is not available, however, we were able to recover the PDS sequence from the available leaf and root transcriptomes (NCBI Bioproject ID No. PRJNA67621).
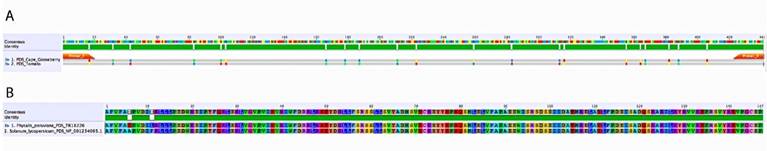
Using the PDS gene from Solanum lycopersicum as a query against the cape gooseberry leaf transcriptome, a fragment with a length of 2 400 bp was detected as the best alignment hit. The VIGS-Tool [27] selected a DNA region between 1 600 bp and 2 100 bp for this putative PDS gene. The amplified fragment had a size of 440 pb and was inserted in the vector pTRV2 (PYL156). The total size of the vector was 11 821 bp (Fig. 1). The fragment sequence was verified by Sanger sequencing and showed a 95 % similarity with 20 bp out of 441 from the S. lycopersicum PDS gene (Fig. 2a). At the protein level, only two amino acid differences out of 147 were found (Fig. 2b), suggesting a highly steady structure of the PDS gene between these two species. The high homology gave us confidence to continue the experiment because several studies have already silenced this gene in tomato [14, 32,33].
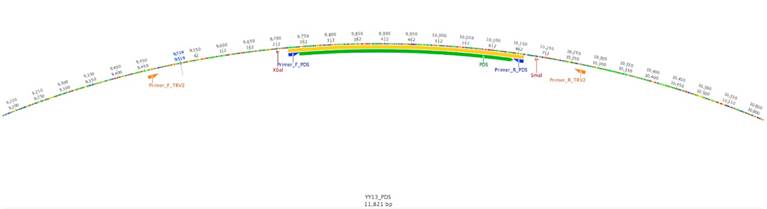
Figure 1 TRV2 vector with the PDS insert. The map was constructed using Geneious software, version 8
Gene silencing experiment
Cai et al. 2006 reported that in Arabidopsis thaliana, the strains C58C1 and LBA4404 were not efficient in silencing the PDS gene. In contrast, the strain GV3101 caused a high efficient silencing in Arabidopsis [24]. TRV vector has been used in many dicot plants to silence genes, and also in several Solanaceous plant species such as tobacco (Nicotiana benthamiana) [25, 30] tomato (Solanum lycopersicum) [30], pepper (Capsicum annuum) [34], potato (Solanum tuberosum) [35], and petunia (Petunia hybrida) [36].
Photoperiod and seedling age were tested for P. peruviana. At first, we chose plants with two fully expanded cotyledons to infiltrate; however, these plants died after a few days, probably due to a defense mechanism response to Agrobacterium. However, in a new assay using plants with three or four true leaves the PDS gene silencing was successful (Fig. 3).
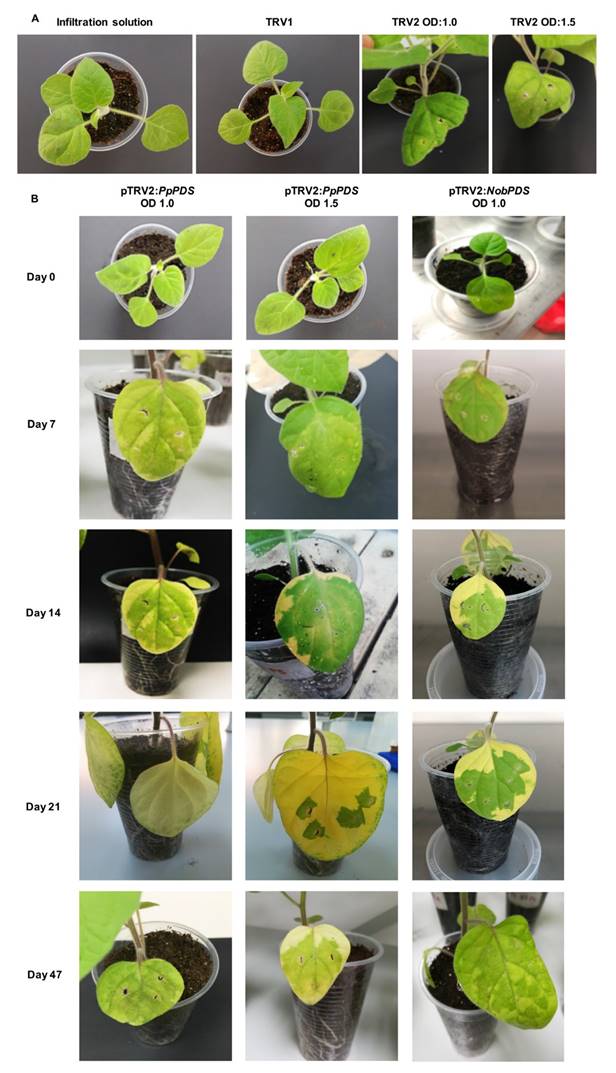
Figure 3 Images of the effect of VIGS via agro-infiltration of GV3101 strain in plants of P. peruviana. a). Control treatments and viral symptoms of GV3101 strains on OD = 1.0 and OD = 1.5 treatments, b). Silencing effects of OD:1.0 and OD:1.5 at different days post inoculation.
The PTGS in plants is well known for causing a defense mechanism response against viral infections [21]. As expected, the negative controls did not show signs of photo-bleaching on the leaves (Fig. 3a); but we observed strong viral symptoms such as changes in leaf form and local necrotic lesions for the two treatments with pTRV2:PpPDS (Fig. 3a). The infection did not seem to have affected other morphological characteristics of the plant.
Gene silencing in gooseberry
Pang et al. 2013 [37] reported 100% photo-bleaching in Gossypium barbadense, known as extra-long staple cotton, using a photo-period of 16 h light/8 h dark, and 92 % using a photo-period of 12 h light/12 h dark after two weeks post-inoculation. The appearance of the photo-bleached phenotype is facilitated under high light intensity and long-day photo-period conditions. We previously tested a photoperiod of 12 h light/12 h dark, in which very small bleach spots across the leaves appeared (data not shown). Despite using 16 h light/8 h dark the photo-bleached phenotype emerged in new leaves 7 dpi and occurred for at least 4 weeks. Afterwards, plants began to recover their normal appearance after 28 dpi; and the symptoms continued to dissappear until the end of the experiment at 47 dpi (Fig. 3b). The recovering process can be attributed to the transient nature of the VIGS. The photo-bleaching phenotype was also observed with the positive control vector constructed for N. obstusifolia. The silenced phenotype was uniformly spread across all the entire leaf in the treatment with OD:1.0 after 21 dpi. In treatment with OD:1.5 and with the pTRV2:NobPDS vector, some patchy patterns were observed (Fig. 3b). Similar results in photo-bleaching time and efficiency have been reported in tomato [30], and recently in Solanum rostratum [38], maize and wheat [39].
The PDS silenced plants showed a patchy photo-bleaching pattern, similar to the one reported by Meng et al. 2016 [38]. We observed some variation in gene silencing between plants, probably due to differences in TRV constructs uptake after agro-infiltration. Besides, Kirigia et al. 2014 [40] reported that the translocation of PTGS silencing factor could be affected because of short-range cell-to-cell movements through plasmodesmata and phloem-associated long-range transport mechanisms. Therefore, further studies in cell physiology in cape gooseberry must be conducted to confirm if this can affect the movement of the vector inside the plant. We found that the concentration of A. tumefaciens in the culture affects gene silencing; cultures resuspended at OD:1.0 exhibited stronger silencing than OD:1.5 (Fig. 3b). This result is probably due to the thick leaves of cape gooseberry, and therefore, less concentrated inoculum can enter more easily by agro-infiltration with a syringe. To increase the efficiency of the infiltration, vacuum infiltration of a culture with an OD of 0.5 reported in other Solanaceous species was tested [38, 41], but no successful silencing effect was observed (data not shown).
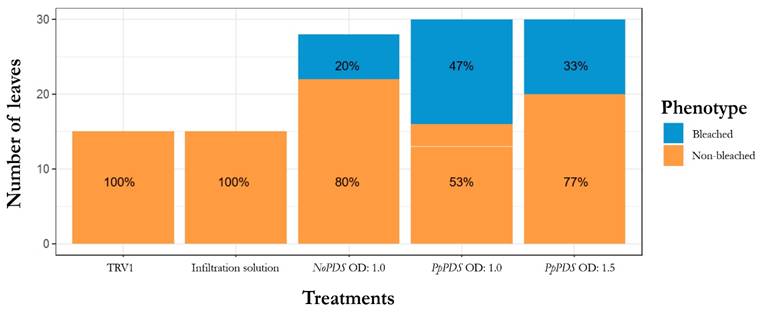
From the 20 inoculated plants with the pTRV2:PpPDS vector, 10 plants showed photo-bleaching leaves indicating a rate efficiency of 50 %, which is consistent with other reports [40, 42, 43] (Fig. 4). For each concentration, the efficiency was the same, however, the plants treated with OD:1.5 showed a patchy photo bleaching, meanwhile leaves of plants treated with OD:1.0 were totally bleached (Fig. 3b). Besides, we observed a higher percentage of bleached leaves in the pTRV2-PpPDS with OD:1.0 compared to the OD:1.5 treatment (47 % and 33 %, respectively) (Fig. 5)
It is well known that VIGS efficacy depends on the plant-virus interactions, which can be influenced by environmental factors and the development stage of the plant [23]. In addition, the length of the vector can also affect VIGS [32]. Further work to test the influence of the environment and length of the insert in VIGS of cape gooseberry may be necessary. It is also critical to assess different plant developmental stages as we mentioned above.
RT-PCR and qPCR analysis
The abundance of PpPDS transcript levels through semi-quantitative RT-PCR was confirmed using primers that annealed outside the targeted region for gene silencing and near the untranslated region (UTR) of the PDS gene. These primers were specifically designed to anneal the plant gene in order to avoid the amplification of the pTRV2:PpPDS construct. As expected, these primers weakly amplified a 176 bp fragment, indicating a low expression of the PDS gene in treated plants (Line 3 and 4) (Fig. 6). In contrast, PDS was highly expressed in control plants (Line 1 and 2), in which no down-regulation was expected (Fig. 6). The amplification of the constitutive gene a-tubulin gave a 133 bp fragment, corresponding to the expected size in all samples.
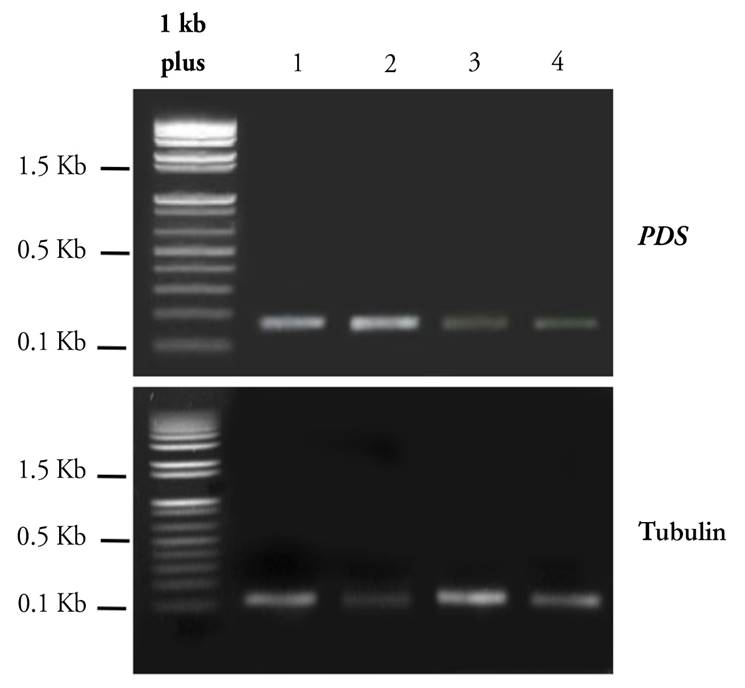
Figure 6 Expression of PDS and α-tubulin by semiquantitative RT-PCR. Lane 1 corresponds to leaf tissue inoculated with infiltration solution, lane 2 corresponds to pTRV1, lane 3 corresponds to pTRV2:PpPDS OD:1.0, lane 4 corresponds to pTRV2:PpPDS OD:1.5. α-tubulin was selected as an endogenous control.
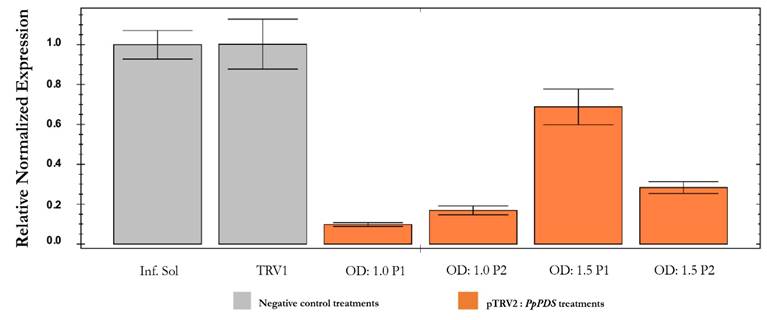
The relative transcript abundance of the PpPDS gene estimated by qPCR was also significantly higher in infiltration solution and pTRV1 treatments than in plants inoculated with the pTRV2:PpPDS construct at different OD concentrations (Fig. 7). As expected, the gene was less expressed in plants with OD:1.0 treatment compared to plants with OD:1.5 treatment, corroborating the phenotypic characterization. As explained earlier, the infiltration process is facilitated with lower ODs. The reduction in pTRV2:PpPDS transcripts in infected plants with OD:1.0 and OD:1.5 was 88 % and 47.5 % respectively, compared to control plants. Hence, the inoculum concentration has a significant influence on the VIGS effectiveness. In addition, the melting curves showed a specific amplification of the PDS and α-tubulin showing two separate peaks with sequence specific shapes at the dissociation curve.
Conclusions
In this study, we established a TRV-based VIGS system that can successfully induce photo-bleaching by silencing the PDS gene in cape gooseberry leaves. Using the Agrobacterium strain GV3101 with an OD of 1.0 and the leaf syringe-infiltration method, silencing efficiency can reach to approximately 50 %. These results constitute the first functional gene study in P. peruviana showing that VIGS using TRV can be effectively used in the future to corroborate the function of candidate genes related to important agronomic traits such as resistance response and fruit quality.