Introduction
The main components of cow milk are water, proteins, fat, carbohydrates, and minerals. These components exhibit quantitative variation both within and between individuals and their proportions in milk chiefly depend on cow race, feed, age, lactation period, time of the year, and milking system [1].
A detection method of bacterias in milk
Along its production chain, cow milk quality and safety might be threatened by biological, physical, and chemical hazards. Inadequate milking and manipulation practices, lack of boiling, and insufficient cooling methods during raw milk harvesting may lead to microbial growth in a very short period, thus increasing the risk of food poisoning and foodborne infections in consumers [2].
The most important biological contaminants of raw cow milk include bacteria like Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes [3]. These bacteria have been associated to food poisoning cases in the United States [4]. In 2014, a total of 140 food poisoning outbreaks were associated to Salmonella, 23 to Shiga toxin-producing E. coli, and 9 to L. monocytogenes; 80 out of the 140 Salmonella outbreaks were originated by consumption of food of animal origin. Among these, 15 cases were associated to the intake of unpasteurized dairy products, 3 cases due to consumption of pasteurized milk products, and 1 case involved a dairy product lacking information on hygienic protocols [4]. In Colombia, various investigations have demonstrated the presence ofpathogenic microorganisms such as Brucella spp., L. monocytogenes, and Staphylococcus aureus in cow milk, and public health surveillance bodies have reported outbreaks of E. coli and coagulase-positive Staphylococcus associated with milk consumption [5, 6].
Commonly employed microbiological identification methods for Salmonella enterica, E. coli O157:H7, and L. monocytogenes in milk and dairy products rely on growing samples in selective media [7, 8]. Depending on the culture medium used, presence/absence of colonies is evaluated. Then, biochemical phenotyping and serotyping assays are performed. These culture-based microbiological detection methods take several days to identifying a given pathogen. In industry and public health surveillance setups, efficient and accurate testing methodologies are needed to ensure the timely release of consumer-safe milk and dairy products into the market. Molecular techniques are fast and efficient to detect pathogenic bacteria in food, particularly in milk. By combining both methodologies, the milk industry can have reliable test results over a short time period [7-10].
A promising molecular method for food testing is the multiplex PCR (mPCR). This method uses a group of target-specific primers to amplify different DNA fragments in a single reaction. With this technique, multiple microbial species can be simultaneously detected since the pool of primers consists of sequences targeted to a given bacterial species [11, 12]. This approach has been successfully applied to detect S. enterica, E. coli O157, and L. monocytogenes in milk [13], meat [14], cereals [15], and kimchi [16], offering the possibility to implement it as a routine technique.
In the present study we assessed the effectiveness of the mPCR technique in detecting S. enterica, verotoxin-producing E. coli (E. coli O157:H7), and L. monocytogenes in experimentally contaminated cow milk. In parallel we conducted culture-based conventional microbiological identification tests, evaluating the use of a culture medium that favors the growth of the three focal bacteria.
Materials and methods
Bacterial strains
The bacterial strains used in this study were obtained from different reference collections. The S. enterica and L. monocytogenes reference strains were obtained from the American Type Culture Collection (strain codes ATCC13076 and ATCC19115, respectively; Microbiologies®, USA) and the E. coli O157:H7 strain was obtained from the Collection of Microorganisms at Universidad Javeriana (strain code CMDM0218; Colombia). In addition, 43 bacterial strains, across 11 genera, were used to assess the mPCR assay specificity (Table 1). These microorganisms were considered because they are also common contaminants of milk [5, 6, 17].
Table 1 Bacterial strains used to assess multiplex PCR assay specificity. 1. Collection Universidad Javeriana; 2. Bank of Germplasm, Bacteria and Virus (agrosavia); 3. Collection Universidad Colegio Mayor de Cundinamarca; 4. INVIMA; 5. Instituto Colombiano Agropecuario (ICA); 6. Universidad Nacional; 7. Work collection agrosavia.
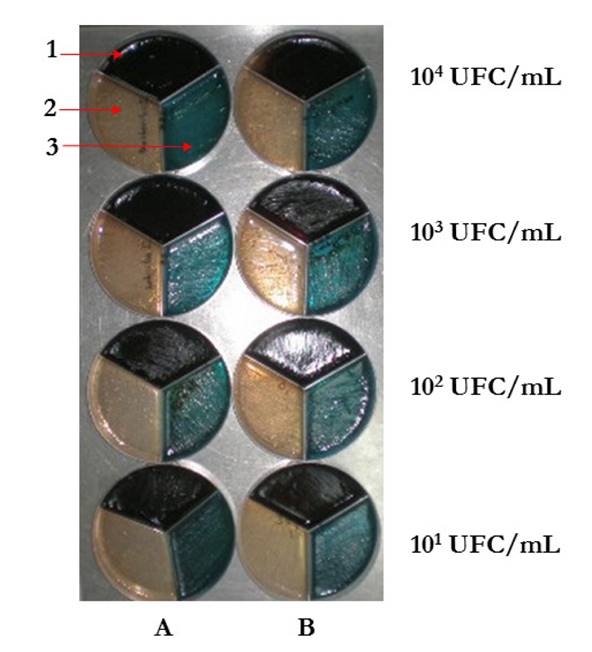
Culture-based approach: simultaneous detection of Salmonella enterica, Escherichia coli O157:H7 and Listeria monocytogenes with SEL broth.
For the simultaneous detection of S. enterica, E. coli O157:H7, and L. monocytogenes in cow milk, experimentally contaminated milk samples with these three microbes were cultured in SEL broth, as a pre-enrichment step, according to the method by Kim & Bhunia [18]. Briefly, seven different microbial concentrations in a range of 100 to 106 UFC/mL, of each microorganism, were used to experimentally contaminate 10 ml of commercial ultra-pasteurized (UHT) milk, which was previously confirmed free of pathogens by conventional microbiological methods and PCR. Each 10 ml contaminated milk sample was used to inoculate 90 mL of SEL broth. SEL broth cultures were incubated for 6 hours at 35 ± 2 ◦C. Later, each dilution was streaked in Hektoen agar, Mac Conkey sorbitol agar supplemented with cefixime and tellurite, and oxford agar [19-22]. All the plates were incubated at 37 ◦C for 24 hours.
In parallel, the set of experimentally contaminated milk samples were evaluated for microbial contamination in three different selective culture media according to FDA and INVIMA recommendations [19-22]: Lactose pre-enriched broth was used to grow Salmonella [21], EC broth supplemented with Novobiocin (EC-N) for E. coli O157:H7, and buffered Listeria medium (Oxoid® Thermo Fisher Scientific, USA) for Listeria. All cultures were incubated at 35 ± 2 ◦ C for 24 hours. After incubation, an aliquot of each broth was streaked in selective agar broths: Hektoen agar, Mac Conkey sorbitol agar supplemented with cefixime and tellurite, and oxford agar. All the plates were incubated at 37 ◦ C for 24 hours.
Molecular approach: simultaneous detection of S. enterica, E. coli O157:H7 and L. monocytogenes by mPCR
The employed DNA extraction protocol followed molecular methods for microbial food contaminants [23-25]. After pre-enrichment in SEL broth, three 1 mL aliquots were taken from each culture; then, bacterial cells pellets were obtained with centrifugation at 4 600 rpm for 5 minutes, followed by two washes with PBS buffer (137 mM NaCl; 2.7 mM KCl; 10 mM NaH2PO4; 2 mM KH2PO4, pH 7.4) with centrifugation at 4 600 rpm for 5 minutes each. Bacterial pellets were re-suspended in 100 pL Tris-EDTA lysis buffer (Tris 10 mM, EDTA 1 mM, pH 8.0; Buffer TE) plus lysozyme (20 mg/mL) and incubated at 37 ◦C for 30 minutes. Then, 467 pL of TE buffer, 30 pL of 10 % SDS, and 3 p L of proteinase K (20 mg/mL) were added to each sample and the mix was incubated at 65 ◦ C for 60 minutes. Next, 100 p L of NaCl 5M and 80 p L of Hexadecyltrimethylammonium bromide (CTAB/NaCl) were added and incubated at 65 ◦C for 20 minutes. A volume of 700 pL of chloroformisoamyl alcohol (24:1) was then added to each sample. Samples were centrifuged at 9 000 rpm at 4 ◦ C for 10 minutes and the resulting aqueous phase retained. A second extraction was completed adding 800 pL of phenol: chloroform: isoamyl alcohol (25:24:1) to the aqueous phase and spinning at 9 000 rpm at 4 ◦ C for 2 minutes. Finally, DNA was precipitated with isopropanol and washed with 100 jL of70 % ethylic alcohol and centrifuging at 4 250 rpm at 4 ◦ C for 2 minutes. DNA samples were dried at room temperature and re-suspended in 50 j L of buffer TE and stored refrigerated until further use.
Multiplex PCR reactions were conducted at a final volume of 25 j l, containing PCR Buffer 1X, MgCl2 1.5 mM, dNTP 0.2 mM. 1.5 U of Taq Platinum® (Invitrogen, USA), and 5 jl DNA (~100 ng of DNA). The set of primers used in each reaction targeted highly conserved genes of S. enterica (gene hisJ) [26, 27], L. monocytogenes (gene hlyA) [25, 28, 29], and E. coli O157:H7 (genes vt1 and vt2) [30]. Primers targeting S. enterica and E. coli O157:H7 genes were used at a final concentration of 5 pmol, and primers targeting the L. monocytogenes gene were used at a concentration of 40 pmol. All primer sequences are shown in Table 2.
Table 2 Primer sequences used for the multiplex PCR (mPCR) detection of Salmonella enterica, Escherichia coli O157:H7 and Listeria monocytogenes.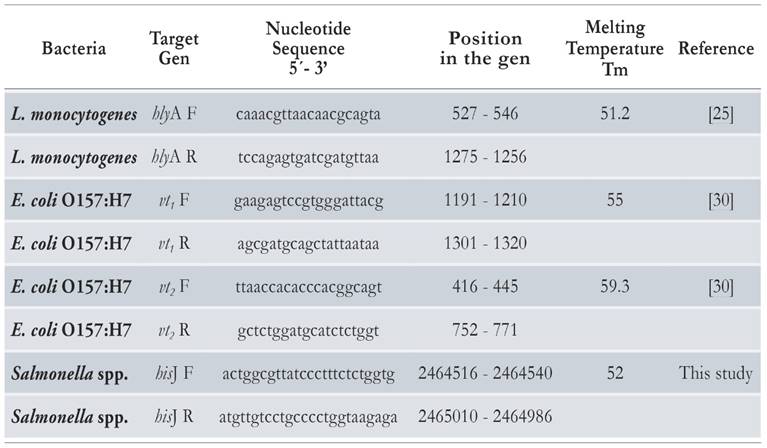
The amplification program consisted of an initial denaturation step for 1 minute at 95 ◦C, followed by 30 cycles of 30 seconds at 94 ◦C, 30 seconds at 50.4 ◦C, and 1 minute at 74 ◦C. The program was completed with one final extension step for 5 min at 74 ◦C. DNA extracted from single reference strain cultures was used as positive control in every mPCR reaction. All mPCR products were visualized in 2% agarose gels in 1X TAE buffer (40 mM Tris-acetate, 1 mM EDTA pH 8 ± 0.2) and stained with SYBR® Safe - DNA Gel Stain (Thermo Fisher Scientific, USA). Electrophoreses were run at 90V for 90 minutes.
The sensitivity of the mPCR was assessed using DNA extracted from milk inoculated with the six concentrations of S. enterica, E. coli O157:H7, and L. monocytogenes. The specificity of the mPCR assay was assessed with DNA isolated from the 43 bacterial strains other than the focal bacterial trio (Table 1). These bacteria were grown in media without selective bacterial inhibitors and expanded in BHI broth, then 1 mL of each culture was used to prepare DNA with a standard phenol-chloroform protocol [24]. DNA extraction of Gram-positive bacteria was performed with lysozyme at a concentration of 2 mg/mL at 37 °C for 30 minutes [24, 25]. Multiplex PCR amplification was performed with primers designed for the selected genes for S. enterica, E. coli O157:H7, and L. monocytogenes strains, as described in the preceding section. A sample of 10 mL UHT milk sample incubated in SEL broth was used as negative control.
Results
Assessment of the SEL broth for the simultaneous detection of S. enterica, E. coli O157:H7, and L. monocytogenes
The SEL broth allowed growth of the three microorganisms in all seven concentrations evaluated. Fig. 1 depicts the growth from the concentrations 104 UFC/mL, 103 UFC/mL, 102 UFC/mL, and 101 UFC/mL in differential selective agars. The detection limit for S. enterica, E. coli O157:H7 and L. monocytogenes in the SEL broth and in conventional broths was 101 UFC/mL showing that this broth allows the growth of the three microorganisms. No bacterial growth was observed in any of the selective broths.
Multiplex PCR sensitivity
The mPCR assay protocol led to successful, simultaneous amplification of the fragments diagnostic of S. enterica (hisJ), E. coli O157:H7 (vt1 and vt2) and L. monocytogenes (hlyA). With this approach, we detected S. enterica and L. monocytogenes from contaminated milk samples and grown in SEL broth at a concentration as low as of 101 UFC/mL, for each microorganism and E. coli O157:H7 at a concentration as low as 100 UFC/mL (Fig. 2).
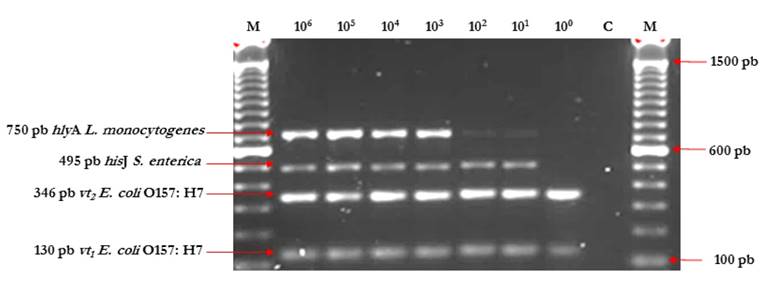
Figure 2 mPCR assay sensitivity results for S. enterica, E. coli O157:H7 and L. monocytogenes, using SEL broth. Lane M: Molecular size marker 100 bp (Invitrogen®). Lanes 1 and 10, M: Molecular size marker 100 bp (Invitrogen®); Lanes 2 to 8 correspond to PCR products from DNA material extracted from experimentallyinoculated milkwithpathogen concentrations: 106, 105, 104, 103, 102, 101, 100 UFC/mL; and Lane 9, C-: Negative control of the PCR reaction.
Multiplex PCR specificity
Fragment amplification with this mPCR assay was strain-specific for L. monocytogenes, S. enterica, and E. coli O157:H7. No amplicons were obtained with any of the DNA material from the additional 43 bacterial strains regardless of their taxonomic proximity to those of the focal trio (Fig. 3). For instance, amplification was negative for strains of other bacterial species in the genera Listeria (Listeria inocua) and Escherichia (non-STEC E. coli).
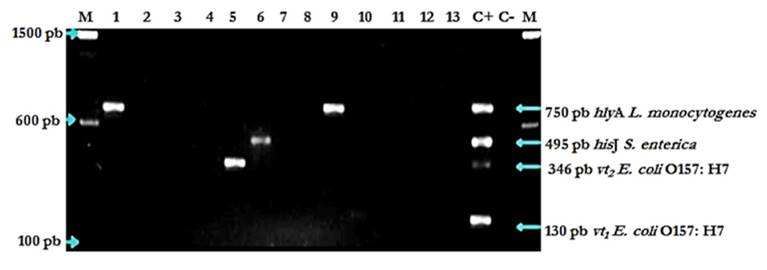
Figure 3 mPCR assay specificity evaluation results. Lanes: 1, L. monocytogenes strain 23; 2, Staphylococcus aureus CMDM080 strain 32; 3, Klebsiella ascorbata CMDM 042 strain 7; 4, Brucella abortus strain 3; 5, Escherichia coli B001 strain 4; 6, Salmonella spp. strain 23; 7, E. coli ATCC 25992 strain 5; 8, Streptococcus spp. strain 38; 9, L- monocytogenes strain 14; 10, Staphylococcus aureus strain 21; 11, Micrococcus luteus strain 19; 12, Pseudomonas sp. Strain 21; 13, Streptococcus agalactiae strain 36; C+: Positive control (S. enterica, E. coli O157:H7, L. monocytogenes); C-: Negative control of the PCR reaction; M: Molecular size marker 100 bp (Invitrogen®).
Discussion
Milk quality and safety are compromised when contaminated with microorganisms such as S. enterica, E. coli O157:H7, and L. monocytogenes. These microorganisms are likely to be present in raw milk and dairy products due to accidental contamination during the milking process or milk handling. These microbial milk contaminants pose a threat to public health if they remain undetected along the path from primary production to the consumer. Time-efficient pathogen detection methods ought to be developed to ensure constant supply of safe foods.
In the present study we evaluated the use of pre-enriched SEL broth to simultaneously detect S. enterica, E. coli O157:H7, and L. monocytogenes in experimentally contaminated cow milk. Overall, bacterial growth in SEL broth was comparable to that achieved in strain-specific pre-enriched broths: lactose broth for S. enterica, EC-Novobiocin broth for E. coli O157:H7, and buffered Listeria broth for L. monocytogenes. Although in the present study the incubation period was shorter, our results agree with those by Kim & Bhunia [18]. Furthermore, growth of the three microorganisms was observed across most of the range of tested inoculum concentrations, per microorganism, (101 to 106 UFC/mL) after a six-hour incubation period at 37 ◦ C. To contribute to the successful detection of several microorganisms in a single mPCR assay it was necessary to use an adequate pre-enrichment step. The universal pre-enrichment broth (UPB; DifcoLab, Sparks, MD®) is commercially available for multiple enrichment of pathogens; however, its lack of selectivity for certain pathogens, renders it unpractical for milk samples with high contamination levels [18]. Consequently, pre-enrichment in SEL broth culture is an efficient way to simultaneously recover the three pathogens in milk samples.
The mPCR assay appeared to be more sensitive in detecting E. coli O157:H7 than the other two focal bacterial strains at their lowest concentration level in contaminated cow milk. However, for abroad range of concentrations, 101 to 106 UFC, the mPCR assay's sensitivity for the three bacterial species was comparable. The mPCR assay was species-specific as revealed by the absence of amplicons when DNA of microorganisms such as Staphylococcus spp., Streptococcus spp. were used as templates. These results showed that this mPCR assay is a feasible technique to detect S. enterica, E. coli O157:H7, and L. monocytogenes in cow milk.
Current research aims at developing highly efficient bulk microbiological food testing methodologies favoring the use mPCR-based approaches [9, 13]. The detection of various pathogens in food via one single assay is an attractive and financially feasible option; it reduces laboratory resources such as space, supplies, reagents, and the work required, optimizing both total cost and running time of the tests [19-22]. The use of a molecular platform for the simultaneous detection of S. enterica, E. coli O157:H7, and L. monocytogenes, with different sensitivity degrees, has been reported for different foods. Using a Real-Time PCR approach, these three microorganisms were detected in milk after an incubation period of 18 hours at 35 ◦C, with a sensitivity of 1 cell/mL, for each microorganism [8, 28, 29]. The same technique has been applied in vegetables with a detection sensitivity of 1-10 cells/mL for Salmonella and E. coli O157:H7 and 1000 cells/mL for L. monocytogenes [13]. Interestingly, in this assay the universal pre-enrichment broth (UPB) was used. This broth was formulated for the simultaneous detection of Salmonella and Listeria in food after incubation at 37 ◦C for 15 hours. The mPCR approach has been used to detect the three microorganisms in other complex food matrixes. For example, in shrimp samples the detection sensitivity was 103 UFC/mL for E. coli O157:H7, 100 cells for L. monocytogenes, and 1-5 cells for Salmonella, using pre-enrichment broth No. 17 incubated for 24 h at 35 °C [9]. In egg samples, the sensitivity was determined at 10 cells/25 g for the three microorganisms, using tryptic soy pre-enrichment broth (TSB) after 15 hours of incubation.
PCR target genes, like specific virulence markers for E. coli O157:H7 and L. monocytogenes have been used to design simple PCR strategies for food and medical sample microbiological analysis. Two targets were chosen for E. coli O157:H7 falling into genes coding for two fractions of the verotoxins (Stx o Vtx). These two genes were chosen because they are highly conserved among enterohemorrhagic E. coli strains which, in turn, are the strains most often implicated in human toxic-infections worldwide. These two E. coli target genes have been extensively studied since 1990 and have been used in developing mPCR techniques for E. coli O157:H7 identification [20, 30].
The pair of primers used for the detection of S. enterica was designed to amplify a histidine transport protein-coding gen which is highly conserved within this genus [26-27]. The listeriolysin O-coding gene hlyA (LLO) is a specific virulence factor for L. monocytogenes, and its amplification in PCR strategies is an excellent target for PCR-detection of L. monocytogenes contamination in meat and dairy products [28, 29].
Conclusions
The method for the simultaneous detection of S. enterica, E. coli O157:H7 and L. monocytogenes in cow milk samples, consisting of pre-enrichment in SEL medium and an mPCR assay is highly specific and sensitive. The A detection method of bacterias in milk specificity of the mPCR assay relied on the choice of four species-specific targets corresponding to the gene hisJ in S. enterica, genes vt 1 and vt 2 in E. coli O157:H7, and gene hlyA in L. monocytogenes. The detection level of this method was 10 UFC/mL for each microorganism. Moreover, this method is likely to be both time- and cost-effective.