Introduction
Persea americana Mill. (avocado) belongs to the Lauraceae family, widely distributed in the world, especially Asia and America. Mexico has at least 26 different avocado-related species (Reyes et al., 2013). At present, three races of avocado are known: Mexican, Persea americana var. drymifolia (Schltdl. & Cham.) S. F. Blake; Guatemalan, P. americana var. guatemalensis (L. O. Williams), and Antillana, P. americana var. americana Miller, which can be differentiated by their morphological-physiological characteristics (Barrientos et al., 2010).
Most commercial avocado varieties are interracial hybrids developed from materials exchanged among areas (Álvarez et al., 2018). In American regions where avocado has been cultivated since pre-Columbian times, production comes from different native trees and select asexually reproduced cultivars. The organoleptic and nutritional characteristics vary according to edaphoclimatic conditions (Mijares-Oviedo & López-López, 1998). They are also a haven of unique genetic diversity for genes that have not yet been studied (Gutiérrez et al., 2015), with native species being the most resistant to pests and diseases (Sánchez, 1999).
Plants are traditionally identified by phenotypic characterization, a procedure that is slow and limited because the expression of quantitative phenotypes is subject to a strong environmental influence (Galindo-Tovar et al., 2008). Molecular markers make it possible to identify, classify, and take advantage of the genetic diversity existing in the genomes of plants since DNA differences or similarities among individuals are observed directly (Galindo-Tovar et al., 2008).
The phylogenetic relationships of avocado have been studied using various molecular markers such as RFLP (Davis et al., 1998; Furnier et al., 1990), minisatellites and microsatellites (Ashworth & Clegg, 2003; Cañas-Gutiérrez et al., 2019a), RAPD (Fiedler et al., 1998; Reyes et al., 2013) with AFLP (Chao, 2003) and SNP (Rendón-Anaya et al., 2019; Talavera et al., 2019). Inter-simple sequence repeats (ISSRs) are highly polymorphic markers and have proven their utility in studies of genetic diversity, phylogeny, gene identification, genetic mapping, and evolutionary biology (Cai et al., 2010).
According to Reyes et al. (2013), Persea indica (L.) Spreng. and P. lingue (Ruiz & Pav.) Nees are not genetically related to the rest of the species studied because they belong to the subgenus Eriodaphne. Moreover, the results allowed differentiating genotypes of the various species of the Persea subgenus, including P. americana and an interspecific hybrid included in the study. However, other studies have been hindered by its genetic complexity and the cross-linked evolution of the species, mainly due to selection and multiple crosses among varieties (Galindo-Tovar et al., 2008). The varied environments in which the avocado has evolved, and how different cultures have managed it, have produced the immense diversity of genotypes currently known (Chen et al., 2009).
These markers have been used in many species, such as species of the genera Agave (Dávila et al., 2007), Plukenetia (Rodríguez et al., 2010), and Manihot (Alzate, 2010).
In Nayarit, Mexico, it is still possible to find wild plants of P. americana that are part of the vegetation, whose characteristics are contrasting, unlike the improved varieties grown intensively in agricultural systems, both in home gardens and backyards (Sánchez, 1999). Avocado growing is economically profitable and represents optimal economic development prospects for the country and especially Nayarit, as it is one of the states with the highest production after Michoacán, Jalisco, and Morelos (Secretaría de Agricultura, Ganadería Desarrollo Rural, Pesca y Alimentación [Sagarpa], 2018). This paper aims to determine the genetic variability of native avocado using ISSR-type molecular markers. The information obtained will help identify and classify the existing diversity and leverage it in genetic improvement programs.
Materials and methods
Genetic material
Ten young leaves were collected from each of 43 avocado trees in the municipalities of Tepic (21°55'60''N, 104°94'70''W), Xalisco (21°42'55''N, 104°89'18''W), San Blas (21°33'56''N, 105°10'59''W), Tecuala (22°39'92''N, 105°44'85''W), and Compostela (21°25'09''N, 21°25'09''N), at an altitude of 950, 1,324, 53, 19, and 190 MASL, respectively, from home gardens, backyards, and wild trees. Besides, a Hass variety of avocado was used as a control (table 1).
Molecular characterization
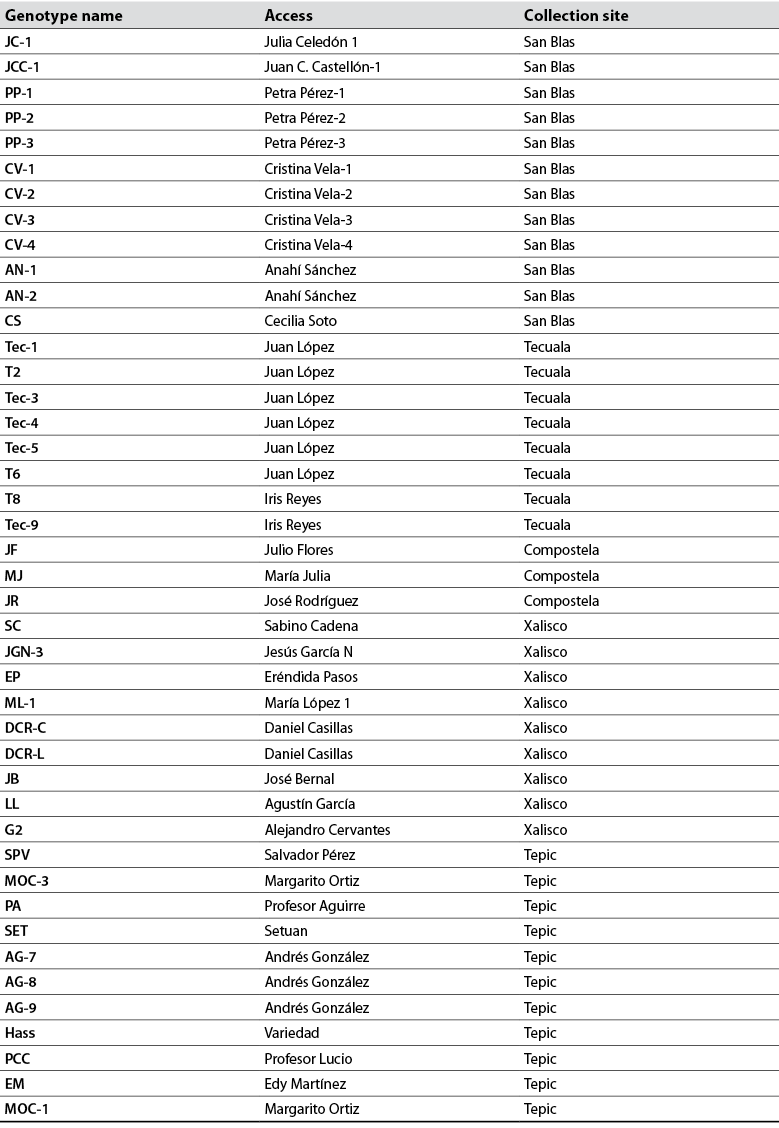
DNA was extracted with 400 mg of lyophilized leaves from each tree, according to the method described by Clarke et al. (1989). The completeness of the extracted DNA was evaluated by electrophoresis in 1 % agarose gels and was in an ultraviolet transilluminator (UVP Benchtop). DNA was quantified in a nanodrop (Jenway, UK). DNA samples were diluted to 10 ng/µL and stored at -20 °C for later use. For DNA amplification, tests were performed with eight ISSR primers reported by Cuiris-Pérez et al. (2009) and used in this study (table 2). The amplification reaction consisted of 4.0 µL genomic DNA (40 ng), 1.5 µL dNTP mix, 2.0 µL ISSR primers (1.0 µM), 2.2 µL 1X Taq buffer, 0.2 µL DNA Taq polymerase (1 U/mL), 0.8 µL MgCl2 (2.0 mM), and 9.3 µL ddH2O. The PCR was carried out in a SelecCycler thermal cycler.
Table 2 List of primers used in molecular analysis and relevant sequences
Source: Adapted from Cuiris-Pérez et al. (2009)
Data analysis
The ISSR data resulted in a binary matrix of ones and zeros for the presence (1) and absence (0) of each band per genotype. The genetic similarity relationship among the studied populations was estimated by the unweighted pair group method with arithmetic mean (UPGMA) and the Dice method. The binary matrix data were analyzed by the Numerical Taxonomic and Multivariate Analysis System (NTSYS) software.
The matrix data for population genetic diversity were analyzed using the TPFGA program (Miller, 1997) and the genetic distances of Nei and Li (1979). The population genetic diversity was quantified according to the number of alleles per locus (A), the percentage of polymorphic loci (P %), observed heterozygosity (Ho), expected heterozygosity (He), the Diversity Index (DI), and AMOVA. An analysis was carried out with TPFGA and GenAlEx (Genetic Analysis in Excel) v. 6.4.
A genetic structure test was performed from an analytical Bayesian approach for assigning individuals, grouping one to five populations. STRUCTURE 2.3.4. (Evanno et al., 2005) was run using 100,000 MCMC (Monte Carlo Markov Chains) iterations, 10,000 replications, and ten repetitions for each K.
Results and discussion
All ISSR primers (Cuiris-Pérez et al., 2009) provided polymorphic information, which allowed characterizing the 43 genotypes under study (figure 1).
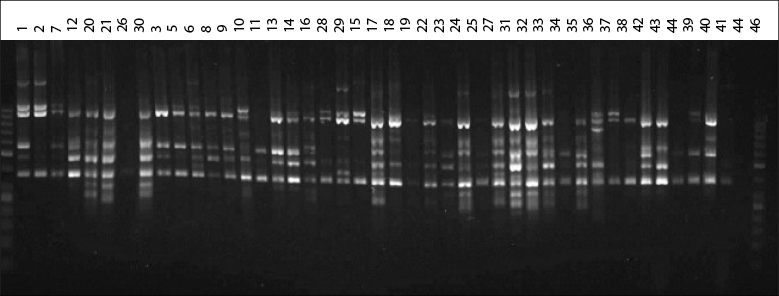
Source: Elaborated by the authors
Figure 1 Genetic profile of native avocado genotypes obtained from the CT8RC primer
Polymorphism level
The eight ISSR-type primers used in 43 native avocado genotypes, belonging to five municipalities, amplified a total of 82 loci. The number of bands detected was from 12 to 17. Other researchers report that the primers used for avocado amplified between 15 and 17 bands and high heterozygosity, like in this study (Galindo et al., 2011). Concerning the information on the polymorphism information content (PIC) detected for each reported sequence, the results indicate that the PIC fluctuated from 36 to 94 % in this research (table 3). Generally, the average PIC was 63.82 % detected with this type of sequence, considered moderate. The results obtained are very similar to the work carried out by Cuiris-Pérez et al. (2009), in which they obtained an average of 92.64 % with 231 genotypes and seven initiators. This type of sequence is considered high, so the ISSRs are suitable for studies of genetic diversity in avocado.
In contrast, Álvarez et al. (2018) reported an average PIC of 31.7 % when using RAPD markers, whereas Bharad et al. (2009) reported up to 73 % and Salazar et al. (2010) 87 % using isoenzymes. Furthermore, the average ID in this work was 64 %, very similar to that observed by Ashworth and Clegg (2003), who reported an average genetic diversity of 60.7 % when analyzing 35 avocado cultivars and two wild relatives with microsatellite markers. However, it is low compared to that observed by Álvarez et al. (2018), who reported values of 84 %.
Table 3 Genetic diversity parameters of native avocado (Persea americana) in five locations sampled in Nayarit, Mexico
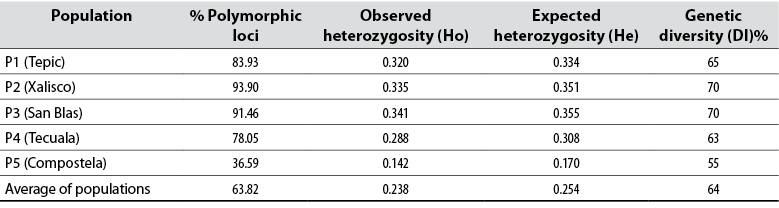
Source: Elaborated by the authors
About He in the different locations, Tepic, Xalisco, San Blas, and Tecuala show very similar values with an average of 0.33, while Compostela had 0.17, which is considered low. The Ho in P3 had an average of 0.34, the highest of all populations, while the lowest value was 0.14, corresponding to P5 (table 3). In a similar work carried out in the inter-Andean valleys of Ecuador, Ponce (2013) obtained Ho of 0.64, while the He was 0.74, which is high. If heterozygosity is considered a measure of genetic diversity, these indices point to moderate genetic diversity in Nayarit, especially when compared with similar works, such as Ashworth and Clegg (2003), who obtained heterozygosity of 0.63.
Table 4 summarizes the data obtained from the AMOVA, which showed that the difference among locations is low (18 %), while the variation within each location (82 %) is broad. Cañas-Gutiérrez et al. (2019a) obtained similar results and showed that most of the genetic variation occurred within the municipality (94.5 %) rather than among municipalities (5.44 %), suggesting that avocados from that part of Colombia are genetically different depending on the sampled site. This finding agrees with the genetic relationships, i.e., the cluster in each node defined by location or climatic condition. The results are due to the genetic differences in the studied avocado collection for materials introduced from other countries and the significant variability of native species. Similarly, based on the AMOVA data, Ponce (2013) obtained 17 % differentiation among provinces and 83 % variation among individuals.
Genetic relationships
According to the cluster analysis of native avocados, well-defined groups were not formed by cross-pollination, but are united by location within groups. Galindo-Tovar et al. (2011) mentioned that, because of the tremendous phenotypic variability and the hybrids among avocado varieties, the identification of genotypes by microsatellites has been ambiguous and makes it challenging to designate variety composition. The resulting dendrogram shows four main groups (figure 2).
Group I comprised the genotypes of five locations: Tecuala (Tec 1, 2, 4, 5, 6, 8, 9), San Blas (CV-3 and 4, JCC-1), Xalisco (SC, DCR- C, and EP), Las Varas (JR, MJ, JF), and Tepic (AG-7 and 8, SPV, PA, MOC-1). Tecuala and San Blas share the altitude (24 to 13 MASL), subtropical climate, and predominance of Solonchak-type soils. Tepic and Xalisco have an altitude between 950 to 1,400 MASL, and Compostela of 190 MASL, with a predominance of Luvisol and Cambisol-type soils, and a tropical climate. We can see that the materials within this group are associated with each other by geographic location. In this sense, Ohsawa and Ida (2007) state that altitude is a reference to estimate genetic diversity since a particular allele shows differences among populations due to natural selection on altitude.
Group II consisted of three locations: Tepic (AG-9, PCC, MOC-3), San Blas (AN-1 and 2, PP-1, 2 and 3, CS, CV-1 and 2) and Xalisco (JGN-3, ML-1, JB, and DCR-L), and Group III of Tepic (SET, EM), San Blas (JC-1) and Xalisco (LL and G2). These two groups are very similar in the location cluster because they have similar altitudes and temperatures, and San Blas is in the coastal area of the state at an altitude of 53 MASL, which agrees with the reports by Cuiris et al. (2009) and Rodríguez et al. (2003) who worked with genetic diversity within a germplasm collection in Cuba and Mexico. These authors comment that, because of the characteristics of ISSRs, they can identify more specific fragments that could share similarities within their genome, regardless of their geographical origin. In Group IV, the Hass genotype was separated from most of the genotypes found in the groups above, except Tec-3, indicating that these two genotypes may be similar. The Hass avocado was used as a reference to observe the genetic difference between a native avocado and a known cultivar.
The results obtained in this research coincide with those presented by Cañas- Gutiérrez et al. (2015). When carrying out the morphological and molecular characterization with AFLP markers of 111 native and commercial avocado (P. americana) cultivars in Colombia, they found high levels of heterozygosity; the genetic distance produced three clusters with individuals very close to each other in similar geographic conditions. Likewise, Cañas-Gutiérrez et al. (2019a, b), using SSR microsatellite markers in native avocados from Colombia, suggest that the structure of the avocado in Antioquia was due to differences in the agro-ecological conditions of the studied area. In their molecular evaluation of native avocado (P. americana) in Nuevo León, Mexico, using RAPD markers, Álvarez et al. (2018) found two large groups that were also associated with each other by their geographical origin, grouping materials from both the northern and southern regions; these regions exhibit varied agro-ecological conditions.
In turn, in a species distinction study of the genus Persea using ISSR, Reyes et al. (2013) grouped some individuals with shared characteristics such as individuals of the Mexican race, individuals of the Guatemalan race, and P. lingue and P. indica of the subgenus Eriodaphne, demonstrating a marked lack of relationship between the species and hybrids of both Persea (except for P. americana races) and Eriodaphne. Talavera et al. (2019) carried out a study in Spain where they used 71 accessions of the three races: Mexican, Guatemalan, and Antillean, using SNP markers. They obtained a low Ho value (0.16) for the molecular marker SSR. These markers widely separated the accessions studied according to the botanical race.
According to Ashworth and Clegg (2003), the clustering conditions of the sampled locations are explained by the complexity of the hybrid state of avocados given the ancient hybridization of the species or a racial differentiation more recent than assumed. Coupled with the old relationship of avocado with humans and various domestication conditions (Galindo-Tovar et al., 2008), it has resulted in a great diversity of genotypes with genetic similarities difficult to separate, shown by the individuals collected for this research. There is some clustering in this study, so Galindo-Tovar et al. (2011) remark that integrating a group of avocados reflects the geographical interaction of four locations in a geographical area defined by similar soil, vegetation, and climate characteristics, but with variations in rainfall and temperature.
The preceding is of particular interest for native avocado in Nayarit, whose marketing problem is the short shelf-life of the fruit; therefore, when genetic variation is found, the possibility of producing new materials increases. Likewise, the characteristic of the native avocados collected could be used for creating varieties with high oil content—higher than that reported for commercial varieties such as Hass—which concurs with research carried out in native varieties collected in Colombia (Álvarez et al., 2018).
The Structure results show four groups (K = 4). In figure 2, each individual is represented by a bar, containing 1 to 4 colors. Each color represents a group, and the highest percentage of color assignment in the bar indicates which group each individual belongs to (figure 2b). Fst 1 of 0.4250, Fst 2 of 0.6948, Fst 3 of 0.4069, and Fst 4 of 0.5547 were calculated for the four groups. The values suggest that the individuals in Groups 1 and 3 are less related to each other than in Groups 2 and 4. These results support the UPGMA tree, showing genetic differences among them for native avocado. Galindo-Tovar et al. (2011) conducted a study of population structure with 60 accessions from seven municipalities in Veracruz, analyzed with SSR microsatellite markers. They identified two groups with an average Fst of 0.2782 and concluded that in both groups, there is little genetic differentiation among individuals according to the Fst values.
Chen et al. (2009), who studied the population structure of central Mexico with wild avocado, found two groups distinguished by elevation and latitude of origin. It is difficult to know precisely to which breed each genotype belongs because of a lack of control of pure breeds and, possibly, the genotypes studied are mixtures of breeds. The authors mention that modern cultivars are complex and inaccurate mixtures of the three races owing to the long period of open pollination and interracial hybridization.
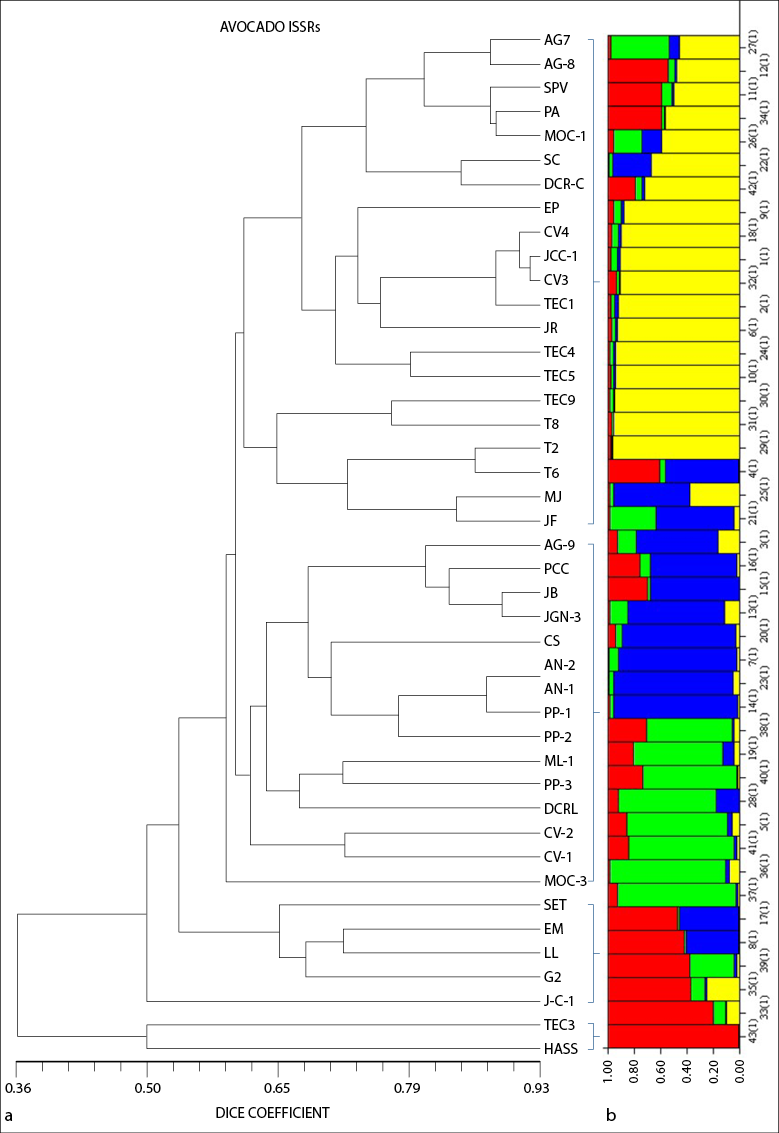
Source: Elaborated by the authors
Figure 2 (a) Dendrogram formed from the Dice distance of the 43 native avocado (Persea americana) genotypes obtained using eight primers (ISSR) and (b) graphic representation of the genetic structure of the six populations where the colors on the X-axis designate the genetic groups (K)
Conclusions
According to the dendrogram, genotypes were grouped by geographical origin and elevation differences, denoting genetic diversity within the same species. All the locations were polymorphic, which shows that native avocados represent a gene pool that could be used for genetic improvement and the creation of new varieties with better commercial characteristics.











 text in
text in