Introduction
In Colombia, potato is one of the most important crops in cold temperature areas (Herrera et al., 2000). However, potato production is affected by Rhizoctonia solani AG-3PT, which is a natural inhabitant of soil (David et al., 2018). AG-3PT is the dominant anastomosis group (AG) most commonly associated with potatoes (Inokuti et al., 2019). Rhizoctonia solani is the causal agent of stem canker and black scurf diseases in Colombian potatoes (Ferrucho et al., 2012), causing necrotic lesions primarily in roots and stems (Aliferis & Jabaji, 2012; David et al., 2018). Consequently, R. solani AG 3PT diseases result in significant yield losses and reduced tuber quality (Beltrán et al., 2007; Muzhinji et al., 2017).
Rhizoctonia solani AG-3PT fungus survives in the soil as mycelia in decomposing tissues and sclerotia on infected tubers and soil (Rioux et al., 2011; Tsror, 2010). Sclerotia adhere to the surface of potato tubers causing black scurf, a severe problem for crop production and distribution and for pathogen-free potato seed production and sale (Inokuti et al., 2019).
The availability of fast and specific diagnostic tests is a pressing need to understand plant pathogens’ ecological, molecular genetics, and epidemiological aspects (Woodhall et al., 2013). The capacity to detect a pathogen and its anastomosis groups in crops would enable researchers to establish its inoculum levels, providing valuable information for decision making during plant disease management (Lees et al., 2002; Woodhall et al., 2013).
Genetic diversity and molecular phylogenetic studies on Rhizoctonia spp. have been limited to the characterization of anastomosis groups and primarily the differentiation between Ceratobasidium spp. and R. solani isolates (Roberts, 2000). Previous research conducted on R. solani had provided evidence of the genetic variability displayed by most anastomosis groups, in some cases suggesting the need to redefine the concept of this species (González-García et al., 2006; Roberts, 2000). Most molecular techniques are based on detecting and genotyping polymorphisms present in the genome at various levels (González-García et al., 2006; Helgason et al., 2003). For example, the analysis of genetic variations in ribosomal sequences helped to identify two phylogenetic groups within AG-3 that have been associated with potato (AG-3PT) and tobacco (AG-3TB) diseases (Kuninaga et al., 2000). Amplified Fragment Length Polymorphisms Analyses (AFLPs) were used to confirm this on isolates obtained from potatoes and tobacco, finding that AG-3PT isolates were not pathogenic to tobacco, and AG-3TB isolates were not pathogenic to potatoes (Ceresini, Shew, Vilgalys, & Cubeta, 2002). Elbakali et al. (2003) characterized R. solani isolates obtained from potatoes in Spain using Internal Transcribed Spacer Regions-Restriction Fragment Length Polymorphism (ITS-RFLP) and Random Amplified Microsatellites (RAMs); AG-3 was differentiated and separated from the other anastomosis groups. However, the RAMs analysis was better suited to genetic diversity studies (Hantula et al., 1996) since ITS-RFLP, which consists of monomorphic band patterns, only allowed the differentiation of the AG under study.
Recently, a genetic structure study on AG-3PT in Colombia showed a low differentiation level between populations, contemporaneous gene flow, and mixed reproduction with evidence of inbreeding, which is possibly associated with high evolutive potential within the studied isolates (Ferrucho et al., 2013). Therefore, it is necessary to understand i) the epidemiological implications of AG-3PT in Colombian potato crops, which are described as the primary etiological agent of the diseases, and ii) the risk level and evolutive potential of the pathogen concerning its genetic diversity.
This study aims to determine the genetic diversity of R. solani AG-3PT present in the Colombian departments of Antioquia, Boyacá, and Cundinamarca, which are important areas in the potato production chain. Furthermore, we evaluated the pathogenicity of R. solani AG-3PT isolates on the potato species Solanum tuberosum and S. phureja, with Phaseolus vulgaris as a non-host control, to observe the range, specialization, and incipient adaptation of the fungus to other species. We concluded that the pathogen R. solani AG-3PT was not host-specific nor genetically isolated and, therefore, it can maintain a wide range of hosts.
Materials and methods
Field sampling and fungal isolation
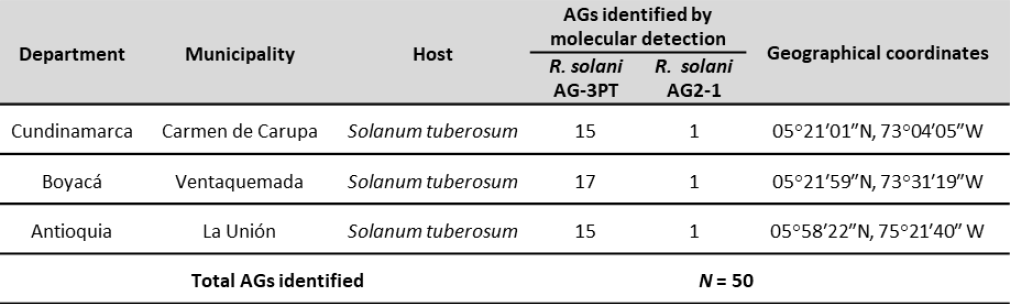
Fifty isolates were obtained from symptomatic tubers on two potatoes cultivars (Diacol Capiro and Parda Pastusa) in Antioquia, Boyacá, and Cundinamarca (figure 1 and table 1). We sampled five rows per field; tuber samples were taken every 10 m from each row following a transept sampling method (McDonald, 1997).
Source: Elaborated by the authors
Figure 1. Origin of Rizocthonia solani isolates obtained from Antioquia, Boyacá, and Cundinamarca in Colombia.
The isolates were in Alkaline Water Agar (AA) (BD Difco™, pH 8.5), and the fungal colonies characteristic of Rhizoctonia spp. were purified using Potato Dextrose Agar (PDA) (BD Difco™) and yeast dextrose- cultured bactopeptone broth (YDB) (BD Difco™), acidified with gallic acid (0.3 g/L) (Beltrán et al., 2007). Sclerotia were transferred to Eppendorf tubes containing silica gel and stored at - 20 °C to preserve the isolates; for non-sclerotic isolates, preservation was done in fungus-colonized rice grains, supplemented with 50 mg/L chloramphenicol (Chavarro-Mesa et al., 2015; Ciampi et al., 2008). The anastomosis or hyphal interaction compatibility tests were performed according to Carling et al. (1998) using UNAL AG-3 as the control strain (this group has the most significant incidence on potato crops in Colombia); it was supplied by the Agricultural Engineering School of Universidad Nacional de Colombia, Bogotá.
Characterization of diametral growth
We evaluated 26 isolates of R. solani AG-3, whose identification codes were RH006, RH015, RH020, RH025, RH027, RH029, RH030, RH033, RH034, RH035, RH040, RH042, RH044, RH054, RH066, RH069, RH075, RH080, RH099, RH106, RH110, RH120, RH140, RH176, RH200, and AG3-Unal. Fivemillimeter discs between 4 to 5 days old were used as inoculum. Discs with the isolates were placed in PDA for incubation at 10, 20, and 25 °C in complete darkness; the assays were performed in triplicate for each isolate. The diametral growth (mm) was measured for the colonies at each temperature after ten days of incubation. Data were analyzed using ANOVA (p ≤ 0.05) and a mean comparison test (Tukey). PAST 3.22 software ran the analysis (Hammer et al., 2001).
Genetic diversity análisis
We extracted the DNA using mycelial growth from PDA cultures, according to Goodwin and Lee (1993). Molecular identification of Rhizoctonia isolates was achieved by PCR (Polymerase Chain Reaction) tests based on selective amplification using primers specifically designed for R. solani AG-3PT: Rs1F2 (5′- TTGGTTGTAGCTGGTCTATTT-3′) and Rs2R1 (5′-TATCACGCTGAGTGGAACCA-3′) (Lees et al., 2002). Genetic diversity analyses were carried out by amplifying the ITS-5.8S rDNA followed by a restriction with the endonucleases AluI, EcoRI (Fermentas®), and TaqI (Promega®). Also, RAMs assay was performed using primers (TCG [5’DHB (TCG)5], GT [5’VHV (GT)7G], and AC [5’DHD (AC)7A]) (Elbakali et al., 2003).
The data obtained here were used to construct a genetic distance matrix based on Nei’s method (Nei, 1973). To visualize the genetic differentiation between the R. solani group AG-3PT isolates, we made a dendrogram and a principal components analysis (PCA) by UPGMA using R programming environment (R Core Team, 2017). The compatibility of the cluster analysis with the original data was evaluated based on the cophenetic correlation coefficient. We assessed the consistency of the dendrogram nodes based on 10,000 bootstraps performed with the pvclust R package (Suzuki & Shimodaira, 2006). The discrimination capacity of the RAMs technique is mathematically defined as the probability that two isolates chosen randomly from a population of nonrelated isolates can be distinguished from one another (Hunter, 1990; Hunter & Gaston, 1988). The reproducibility of the RAMs technique was tested according to Pérez et al. (1998), evaluating the parameters of auto similarity, band repeatability, and the average frequency of each band.
Cross pathogenicity test
We selected four R. solani AG-3 PT isolates from S. tuberosum (RH001T, RH002T, RH004T, RH006T) and four isolates of S. phureja cultivar “yema de huevo” obtained in Cundinamarca in 2018 (RH001P, RH002P, RH004P, and RH005P) to perform cross pathogenicity tests on S. phureja, S. tuberosum, and P. vulgaris. R. solani AG-3PT inoculum was obtained from colonized parboiled rice and attached with parafilm to the base of the last or second-to-last leaf of the main stem in plants with four leaves (15 days after sowing). The inoculated plants were kept in a greenhouse with high humidity (95 %) and daytime temperatures between 15 and 20 °C. An evaluation was performed 15 days after the inoculation by measuring the lesions on leaves and stems. The disease index was calculated according to three severity scales described by Horsfall and Barratt (1945), Jia et al. (2007), and Salazar-Zuluaga (2014).
The experimental design employed was completely randomized blocks with eight treatments and four replicates and one repetition of the entire experiment in time. The mean disease severity for the groups of isolates followed by the same letter (figure 7) is not significantly different according to a contrast of means at p ≤ 0.05 (Chavarro-Mesa et al., 2015, 2020; Ramos-Molina et al., 2016). The rating of the experimental unit involved the combined scores from the four plants. Linear and independent ANOVA models were created for each evaluation parameter (Chavarro-Mesa et al., 2015; Ramos-Molina et al., 2016) using the R programming environment (R Core Team, 2017) to detect evidence of host specialization.
Results and discussion
Fungal isolation and growth characterization
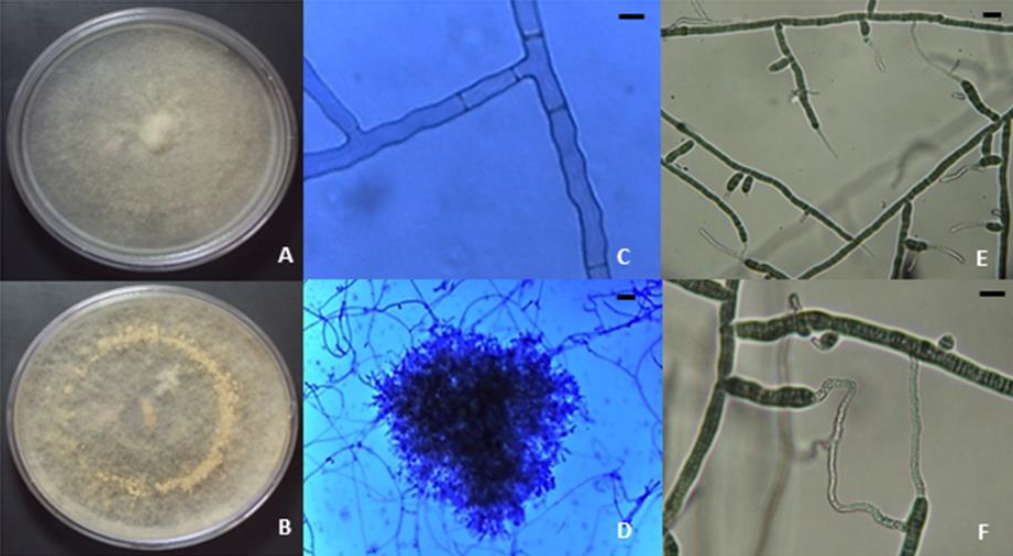
The occurrence of R. solani AG-3PT infection in potato crops found in this study correlates with previous reports in Colombia (Ferrucho et al., 2012) and worldwide (Das et al., 2014; Muzhinji et al., 2015; Tsror, 2010; Yang et al., 2015, 2017). Regarding macroscopic characteristics of R. solani, the isolates developed the usual white/colorless mycelial growth during the early stages, turning light brown after reaching maturity (figures 2A and 2B) (Ceresini et al., 1999). In addition, we observed microscopic characteristics such as 90° angles formed by branching hyphae of septate mycelium (figure 2C) (Ajayi-Oyetunde & Bradley, 2017; Ceresini et al., 1999) and hyphae clumps originating sclerotia (figure 2D) (Ajayi-Oyetunde & Bradley, 2017). Furthermore, the multinucleated state of the fungus was also determined, as was anastomosis type C3 (Carling et al., 1998), because hyphae displayed a perfect fusion between R. solani AG-3PT hyphae (figures 2E and 2F) (Ajayi-Oyetunde & Bradley, 2017).
Photos: Edisson Chavarro-Mesa, Néstor Andrés Herrera-Blanco, Camilo Rubén Beltrán-Acosta
Figure 2. Macro and microscopic characteristics of Rhizoctonia solani. A) R. solani AG2-1; B) R. solani AG3; C) 90° angled branching of septate hyphae, 40x; D) monilloid cells of sclerotia, 10x; E) hyphal anastomosis reaction for the anastomosis group designation of R. solani AG-3 isolates, 40x; F) Hyphal anastomosis and multinucleate R. solani, 40x. Bars = 10 µm.
The AG-3 group of R. solani was present in the three evaluated regions (figure 1), as observed by Carling et al. (2002), Cedeño et al. (2001), and Ferrucho et al. (2012), who established the AG-3PT group as the most common and widespread and the leading cause of potato rhizoctoniosis in Colombia. Color diversity among the colonies was also noted; they ranged between light yellow, light brown, and dark brown (figure 3B). Additionally, there were differences in the size, shape, and color of the sclerotia and the distribution of the culture medium (figure 3B).
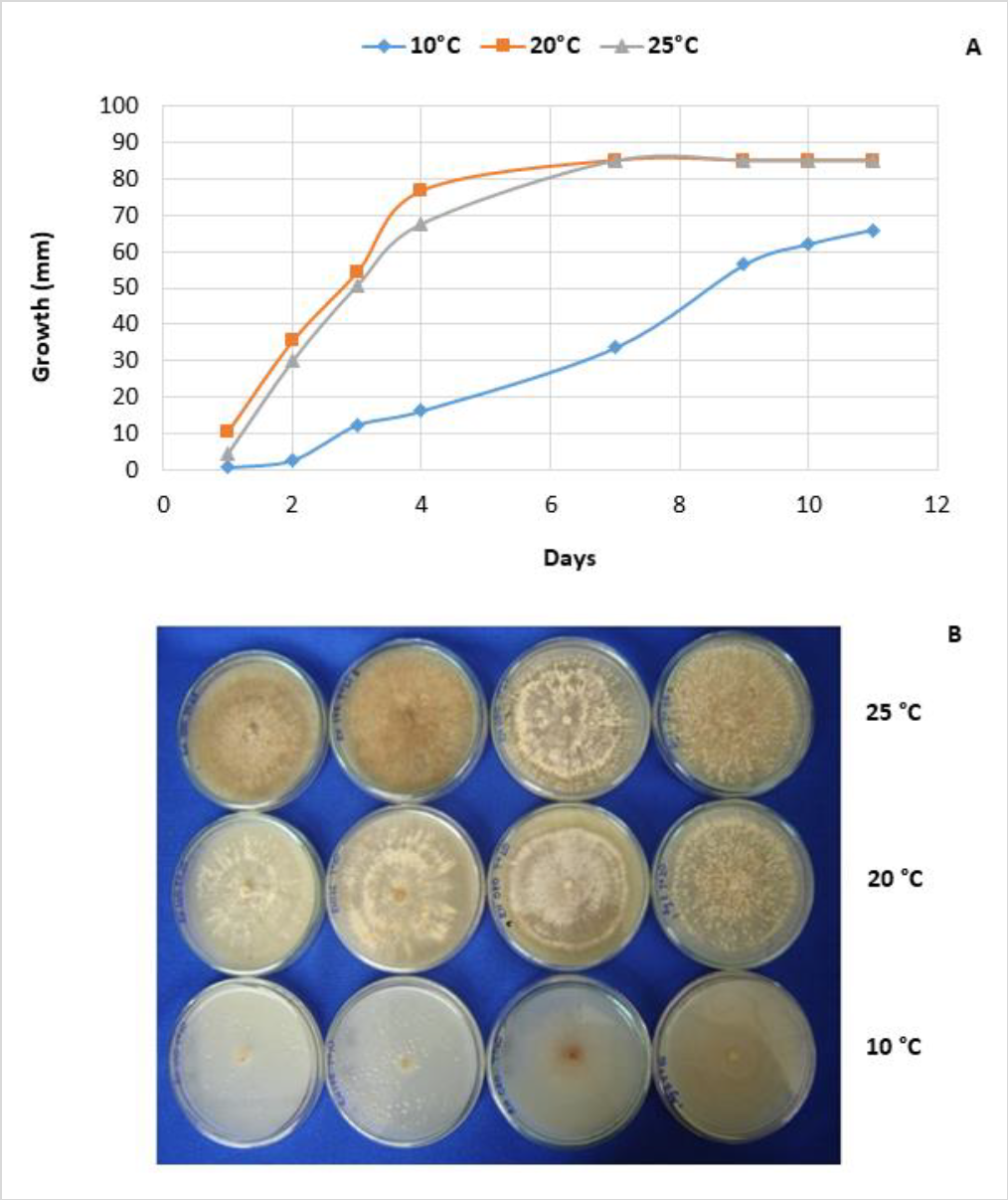
About the growth characterization of R. solani (figure 3A), all isolates showed the highest growth at 20 and 25 °C. On the sixth day, the growth was between 65and 85 mm at 20 °C, and between 73and 85 mm at 25 °C. In contrast, at 10 °C, 75 % of the evaluated isolates showed diameters between 30 and 50 mm after ten days of incubation. The remaining 25 % had colony diameters between 51 and 75 mm (figure 3A). Notice that the development of sclerotia was favored at 25 °C (figure 3B). The presence of AG-3PT is related to environmental conditions such as temperature, prevailing in cold climates (Anguiz & Martin, 1989; Campion et al., 2003).
Source: Elaborated by the authors. Photos taken by Edisson Chavarro-Mesa, Néstor Andrés HerreraBlanco, Camilo Rubén Beltrán-Acosta
Figure 3. Diametral growth of R. solani mycelium AG-3 at 10, 20, and 25 °C. A) Mycelial growth relative to 26 isolations, mean of six repetitions (combination of two independent experiments with three replicates each); B) macroscopic characterization of R. solani at three different temperatures (10, 20, and 25 °C), colonies’ color diversity.
Genetic diversity
The molecular diagnostics methods allowed identifying R. solani AG-3PT and R. solani AG2-1 (figure 1) from the 50 analyzed isolates, which were further differentiated using species-specific markers (Lees et al., 2002; Woodhall et al., 2013). Rhizoctonia solani is a pathogen that can survive on tubers and soil; thus, the detection of inoculum levels on tuber seeds and soil becomes relevant when attempting to predict the potential damage to farming and nonfarming areas, and it is an essential factor in managing the disease (Tsror, 2010). It must be highlighted that the phytopathogenic fungus R. solani is often transmitted through contaminated tubers that are eventually used as seeds, which can establish a long-distance dispersion mechanism and infect pathogen-free regions. Once established in the soil, the mycelia and sclerotia of the microorganism provide a continuous source of primary inoculum (Tsror, 2010).
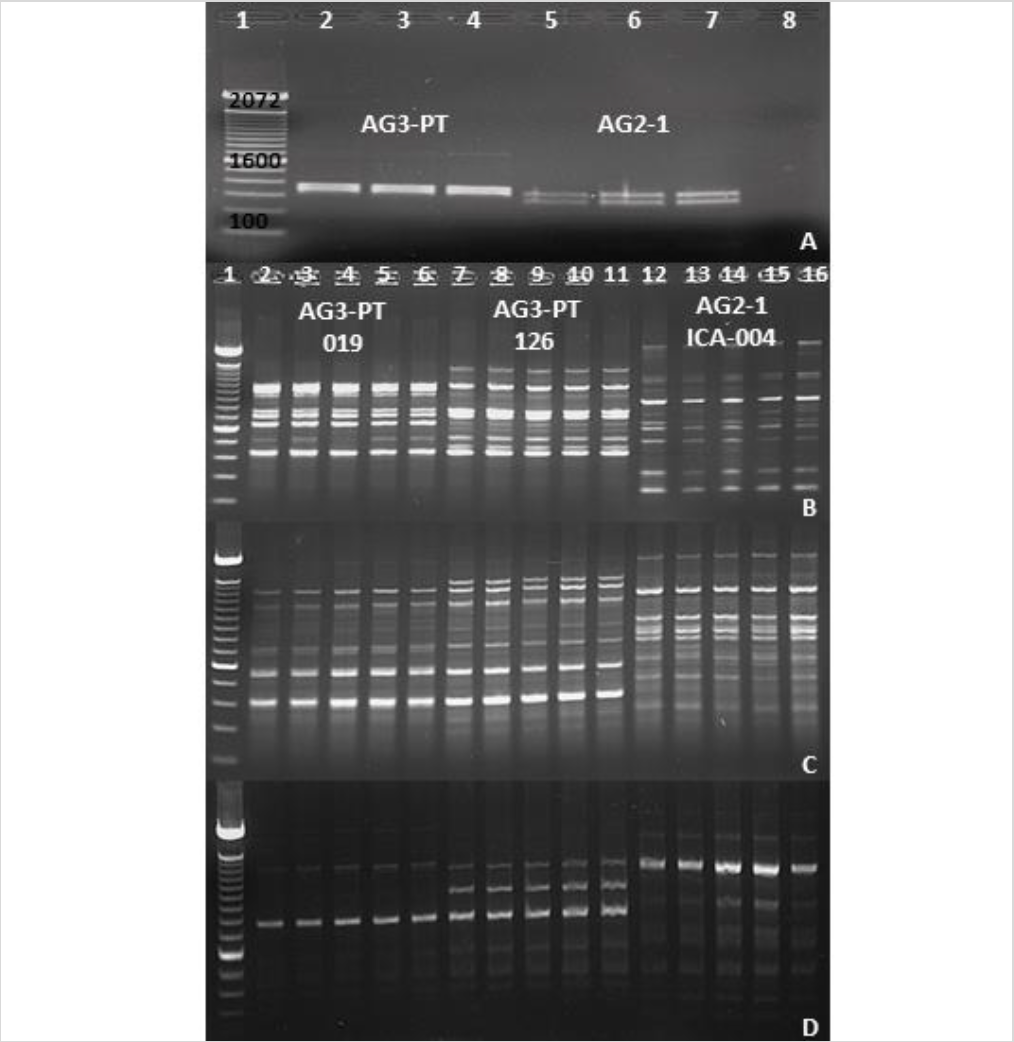
With the development of specific and sensitive techniques, such as PCR and qPCR to detect R. solani AG-3PT, it is now possible to perform epidemiological studies in Colombian potatoes crops (Lees et al., 2002; Woodhall et al., 2013). The restriction analysis of the ITS-5.8S rDNA region using the endonuclease EcoRI in two anastomosis groups (AG-3PT and AG2-1) allowed the specific identification of each group from isolates collected in Antioquia, Boyacá, and Cundinamarca, revealing different band patterns between AG-3PT and AG2-1 (figure 4A).
Source: Elaborated by the authors. Photos taken by Edisson Chavarro-Mesa, Néstor Andrés HerreraBlanco, Camilo Rubén Beltrán-Acosta
Figure 4. Identification and differentiation of AG-3 PT and AG2-1 using molecular markers. A) Restriction analysis with endonuclease EcoRI targeting the ITS-5.8S region in AG-3PT and AG2-1 isolates. Lane 1, 100 bp molecular weight marker (Invitrogen®); lanes 2-4, R. solani AG-3PT isolates; lanes 5-7, R. solani AG2-1 isolates; lane 8, PCR negative control. B), C), and D) Profile differentiation of AG-3 PT and AG2-1 isolates using AC, GT, and TCG primers (RAMs). Lane 1, 100 bp molecular weight marker (Invitrogen®); lanes 2-6, five replicates of the AG-3PT isolate (019); lanes 7-11, five replicates of the AG-3PT isolate (126); lanes 12-16, five replicates of the AG2-1 isolate (ICA 004). Positive and negative controls were included in each run of PCR-RAMs reactions.
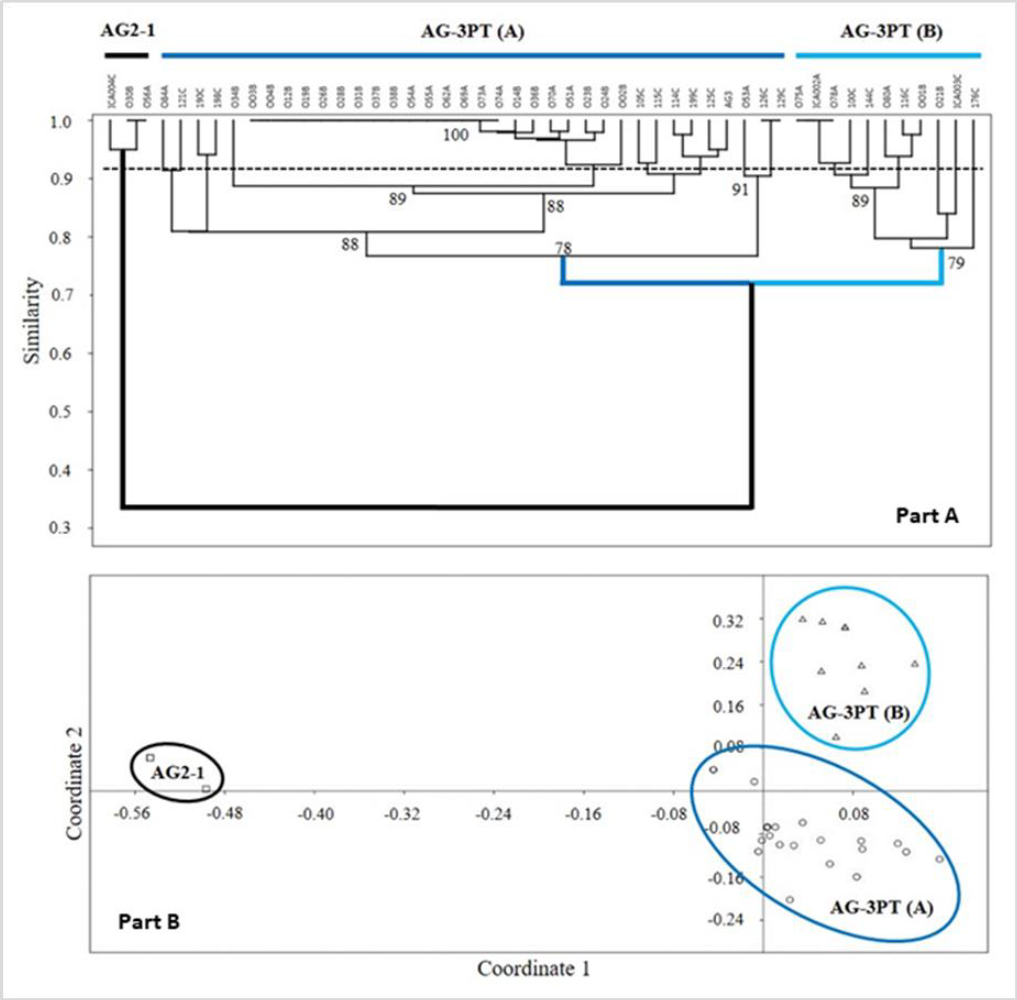
The genetic diversity of the R. solani AG-3PT isolates also revealed polymorphic RAMs band patterns (figures 4B to 4D). The resulting dendrograms (figure 4A) showed two R. solani AG-3PT subgroups that (A) share a similarity index of 78 % in comparison to AG-3PT, and (B) bear a similarity of 79 % among isolates. Besides, the isolates 056A and ICA 004C (AG2-1) showed a similarity of 34 % when compared to the AG-3PT isolates (figures 5A and 5B). The primers AC and GT produced a single monomorphic band in all isolates, resulting in 92.3 and 91.6 % polymorphic loci. These values were higher than those obtained with the TCG primer, which resulted in only three polymorphic bands (42.8 % of polymorphic loci). The average genetic diversity values of R. solani AG-3PT were estimated to be 0.15, 0.27, and 0.31 for each primer set (TCG, AC, and GT, respectively); in turn, the global average (Hj) for Nei’s genetic diversity was estimated to be 0.25.
Furthermore, the isolate groups evaluated through R. solani AG-3PT dendrograms using RAMs did not show any geographical relation between them (figures 5A and 5B). The latest suggests a clonal expansion and dispersion of evolutionarily adapted clones within R. solani AG-3PT populations. Previously, Ceresini, Shew, Vilgalys, Rosewich, & Cubeta (2002) found evidence of R. solani AG-3PT clonal isolation and recombination in North Carolina, USA, through AFLP analysis. Additionally, Justesen et al. (2003) used ITS sequencing and random amplified polymorphic DNA (RAPD) analyses to find clonal isolates of AG-3PT in Danish potato crops. Other studies consider this clonal trait a result of its evolutive potential (Mandujano et al., 2001; Mogie & Hutchings, 1990). Interestingly, this clonal capacity can be observed as an ecological alternative that enables the fungus to favor the dispersion of a specific genotype through space and time (Mandujano et al., 1998, 2001).
Source: Elaborated by the authors
Figure 5. RAMs analysis: Part A) UPGMA similarity tree for the R. solani AG-3PT PCR-RAMs amplicons using standardized Nei’s method; letters followed by the isolation code correspond to A = Antioquia, B = Boyacá, and C = Cundinamarca. Node numbers represent the resampling analysis support value. The horizontal dotted line represents the cutoff estimated for the AG-3PT group isolates. Part B) Principal component analysis based on the similarity indexes of RAMs patterns for R. solani AG-3PT, reconfirming the observations from the dendrogram tree.
Our results revealed similarities of 78 and 100 % between AG-3PT isolates, suggesting that RAMs analysis is a suitable tool for evaluating the genetic diversity of a given population. These markers target sequence changes in microsatellites and can therefore be used to perform a complete genome analysis, facilitating the detection of polymorphisms. Interestingly, the analysis of AG2-1 isolates revealed a similarity percentage of 34 % compared to the entire AG-3PT population. The low similarity value between AGs demonstrates that in comparison with other molecular techniques, RAMs detect genetic dissimilarities between AGs (Elbakali et al., 2003; Justesen et al., 2003) (figures 4B and 5A-B).
Nei’s genetic diversity value observed in this study was 0.25, similar to values from previous reports of 0.28 to 0.38 (Ceresini, Shew, Vilgalys, & Cubeta, 2002) and 0.29 (Justesen et al., 2003). Recently, Muzhinji et al. (2017) reported that South African populations of R. solani AG-3PT are genetically differentiated, with Nei’s genetic diversity values from 0.24 to 0.27. High genetic and genotypic diversity combined with a mixed reproduction capacity and high genetic flow between genotypes within populations are characteristic traits of phytopathogens with high evolutive potential (McDonald & Linde, 2002; Willi et al., 2011).
Recent studies demonstrated that like anastomosis group AG-1 IA, the AG-3PT group possesses a high evolutive potential; this phenomenon requires special attention regarding the proper planning of control strategies and disease management. In addition, the prolonged application of fungicides results in longlasting resistance periods within R. solani populations (Chavarro-Mesa et al., 2015; Ferrucho et al., 2013; Gonzalez-Vera et al., 2010), and therefore, its integral management through biological control strategies could be a viable alternative for the phytosanitary maintenance of potato cultivars in Colombia (Beltrán et al., 2011; Cotes, 2018).
The discriminatory capacity of RAMs in the present study was 94 %, whereas the average auto similarity (Sxx’) and average band repeatability (Rb) values were between 0.96 and 1.00. The average band frequency (fb) showed values between 0.54 and 0.56, probably because the AG-3PT and AG2-1 isolates share few loci (Hunter, 1990; Hunter & Gaston, 1988). RAMs are reproducible over time, and a lack of prior knowledge of the genome under evaluation demonstrates its efficiency at the rapid identification of AGs within the R. solani complex. Also, this same technique can be used in the genetic differentiation of isolates within the same species. AGs have been compared using DNA/DNA hybridization, RFLPs, RAPD, RAMs, isoenzyme electrophoresis, pectic zymograms, and DNA restriction mapping in the rDNA ITS 5.8S region, all of which indicate that the different AGs and subgroups are discrete evolutionary units (Balali et al., 1996; Carling et al., 2002; Elbakali et al., 2003; Kuninaga et al., 2000, 2002; Sharon et al., 2006).
The importance of genetic diversity analyses is directly related to the capacity of a species to survive variations in their environment, primarily because higher genetic diversity represents an increased probability of adapting to these continuous variations. However, species with low genetic diversity face a greater risk against environmental variations, impacting their population and increasing the rate of inbreeding, which further reduces the genetic diversity and makes the species vulnerable to extinction (Ramanatha & Hodgkin, 2002). Concerning phytopathogenic populations, genetic diversity is primarily related to its evolutive potential and population structure. High-risk pathogens that impact agricultural endeavors have high levels of genetic diversity and mixed reproduction systems (sexual/asexual), allowing the emergence and permanence of “new” pathogens in the agroecosystems (Chavarro-Mesa et al., 2015; McDonald & Linde, 2002; Pereira et al., 2016; Willi et al., 2011). Moreover, this pathogen displays a highly conserved evolutive potential subject to the population structure of the fungus observed in Colombia, which could impact the management strategy and alternatives for this disease.
Pathogenicity assays
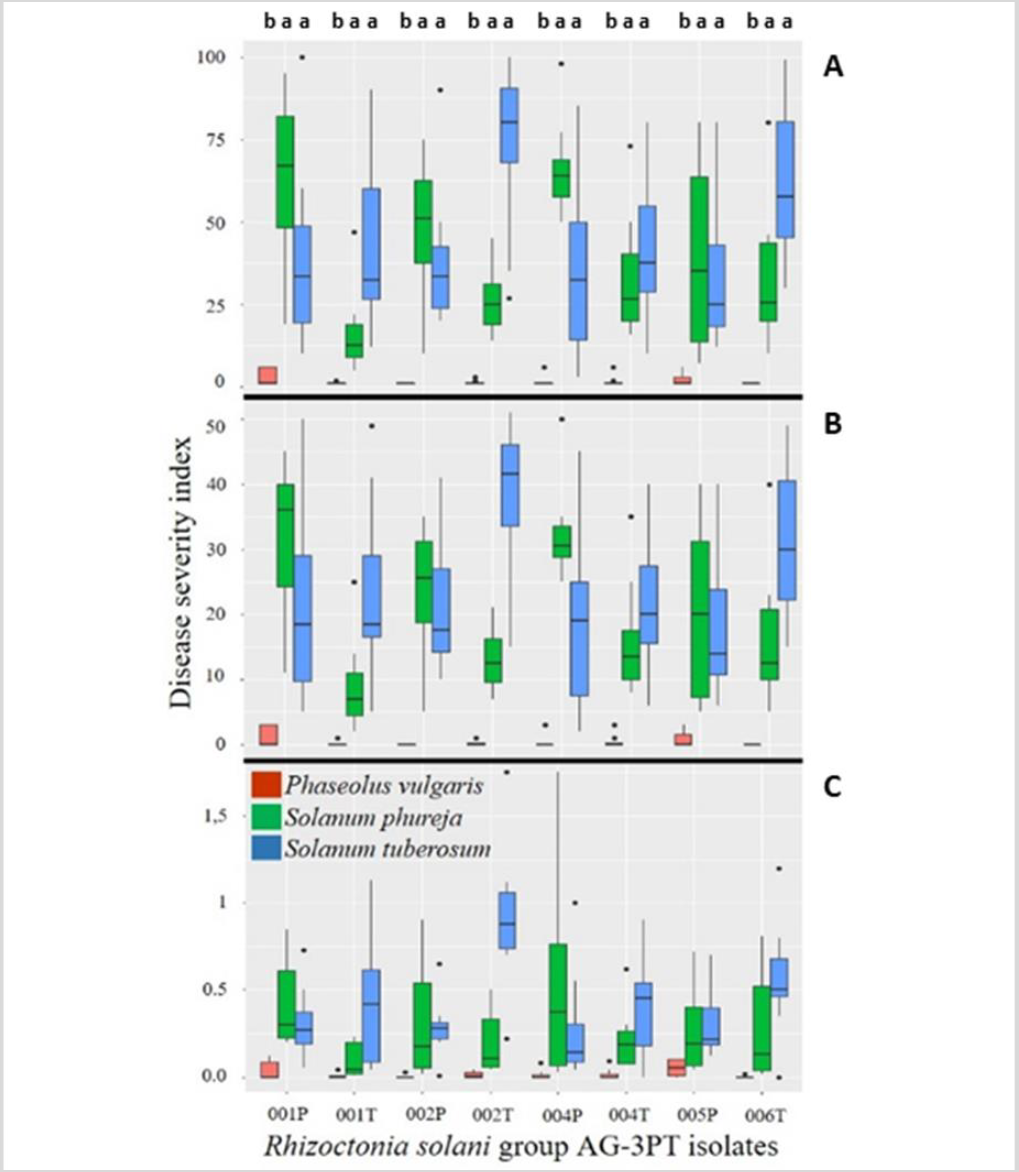
Symptoms present on different hosts (S. tuberosum and S. phureja) corresponded to the common symptoms of R. solani (figures 6A and 6B), such as stem canker and aerial blight of leaves. Pathogenicity was determined by estimating the lesion size on stems and leaves; in some cases, isolates showed distinct host ranges.

Photos: Edisson Chavarro-Mesa, Néstor Andrés Herrera-Blanco, Camilo Rubén Beltrán-Acosta
Figure 6. Pathogenicity test of Rhizoctonia solani group AG-3PT isolates in two different hosts (Solanum phureja and S. tuberosum). A) R. solani group AG-3PT isolates from S. phureja; B) R. solani group AG-3PT isolates from S. tuberosum.
Our results showed that the adaptation of R. solani AG-3PT to S. tuberosum promoted the appearance of symptoms in other potential hosts, such as S. phureja (figure 6). These fungal isolates, which were in the initial stages of specialization, were not yet genetically isolated and therefore maintained a potentially wide range of hosts.
Source: Elaborated by the authors
Figure 7. Cross pathogenicity tests of R. solani anastomosis group-3 PT isolates from S. phureja (RH001P, RH002P, RH004, RH005P) and S. tuberosum (RH001T, RH002T, RH004T, RH006T) in Cundinamarca, Colombia. Three different severity scales were used to evaluate the plant pathogen disease: A) Horsfall & Barratt (1945), B) Salazar-Zuluaga (2014), and C) Jia et al. (2007). Phaseolus vulgaris was not a host of R. solani group-3 PT. S. phureja, S. tuberosum, and P. vulgaris experiments were conducted independently.
Although many R. solani AGs are host-specific, some isolates have shown damage in different plants from their original host (Anderson, 1982; Ogoshi, 1987). Results described by Ferrucho et al. (2012) showed that R. solani AG-3 infected potatoes, tomatoes, and “lulo” or “naranjilla” (Solanum quitoense), exhibiting stem cankers; also, peas, beans, corn, and carrot showed root infection with the fungi. Implementation of crop rotation systems should consider non-hosts of AG-2-1 and AG-3PT. Interspecific adaptation events have been observed in other AGs; recently, it was determined that R. solani AG-1 IA isolates from Urochloa spp. in Colombia had the adaptive potential to become a pathogen in corn. These isolates from signal grass were more aggressive to the same host than the isolates from soybean, suggesting incipient host specialization (Poloni et al., 2016). Another study showed that signal grass isolates of R. solani AG1 IA were more aggressive to Urochloa brizantha “Toledo” than those from rice (Chavarro-Mesa et al., 2015). Finally, R. solani AG-3PT isolates from Cundinamarca feature the adaptive potential to emerge as a pathogen in Andean potatoes (S. phureja) in Colombia, probably because of the similarity between pathosystems (figures 6 and 7).
Conclusions
We isolated and characterized 50 isolates from different crops in three Colombian regions, finding that R. solani AG-3PT was the most frequently fungi associated with potato cultivars. Growth characterization showed that all temperatures were adequate for the growth of the phytopathogen; however, 25 °C seemed to be the most favorable in vitro. Also, our molecular characterization employing different markers reaffirmed that this pathogen has a highly conserved evolutive potential associated with the genetic population structure found in Colombia. In combination with the pathogenicity tests, this characterization indicated the potential of this fungi to be an incipient pathogen in Andean potatoes (S. phureja).