Studies of genetic variation among marine fish populations have allowed aspects of their dispersal and the connectivity among their habitats to be elucidated. Molecular markers are a very important tool for identifying such genetic variability that includes microsatellites, which are sequences of tandem repeats of nuclear DNA that have been widely used for their high degree of polymorphism, Mendelian inheritance and codominance (Selkoe and Toonen, 2006). However, to be used, microsatellites must be isolated, which is a long and expensive process; therefore, for certain investigations, it is often preferable to test primers that have been designed for other, phylogenetically close species because the regions flanking the microsatellites can be highly conserved among species belonging to the same genus or family (Rico et al, 1996; Barbará et al, 2007). This process is called cross-amplification, but it is not always successful because it depends on factors such as the level of polymorphism, the unit of primer repetition and the phylogenetic closeness of the species in which it is implemented. In some cases, specific microsatellites tested in another species of the same genus or family do not present the same level of polymorphism, are monomorphic or are difficult to amplify (Landínez-García et al, 2009; Muñoz et al, 2011; Fresneda -Rodríguez et al., 2013; Almanza et al, 2016).
The genus Haemulon, which belongs to the family Haemulidae, is of great ecological and economic importance in the reefs of the New World (Rocha et al., 2008), and it has been used as a model to analyze evolutionary hypotheses, speciation and diversification in the Caribbean (Rocha et al., 2008, Tavera et al., 2012, 2018). The species of the genus have a wide geographical distribution and inhabit contrasting environments (Tavera et al., 2012). Among haemulids, two representative species are Haemulon aurolineatum and Haemulon steindachneri, which are important targets in artisanal fisheries in the southern Caribbean.
To date, 11 pairs of primers have been designed (Williams et al., 2004) that have been standardized and used in the population genetic analysis of Haemulon flavolineatum (Purcell et al, 2006). The aim of this study was to design primers for eight microsatellite sequences of H. aurolineatum previously registered in GenBank by Turner et al. (2005) and to standardize the amplification conditions of the primers designed by Williams et al. (2004) between H. aurolineatum and H. steindachneri.
Using the microsatellites developed in the present study allows us to answer, with great resolution, questions related to the diversity and genetic structure of these two species, using a more efficient and cost-effective strategy than isolating microsatellites specific to each species.
In April 2016, H. aurolineatum (n = 30) and H. steindachneri (n=30) individuals were collected by artisanal fishermen in Barú-Colombia (10°12'37''N, 75°34'45''W). DNA extraction and purification were carried out using the salting-out extraction protocol proposed by Alhjanabi and Martínez (1997), and DNA quality was evaluated by electrophoresis in 1% agarose gel and quantified by means of a Nanodrop 2000 spectrophotometer (ThermoFisher Scientific, USA).
The primers for the Hau034, Hau075, Hau183, Hau220, Hau315, Hau417, Hau462 and Hau474 sequences were designed with the program Geneious 9.0 (Drummond et al., 2010) (Table 1). In addition to the above oligonucleotides, six pairs of primers designed by Williams et al. (2004) for H. flavolineatum were selected accounting for the level of polymorphism and the composition of the repeat unit. During PCR, the forward primers of each microsatellite were hybridized to the universal M13 sequence with fluorochromes (PET, NED, VIC and FAM) at the 5' end for genotyping (Schuelke, 2000).
Table 1 Characteristics of standardized microsatellites. Rep: Nucleotic repetition; %GC: Guanine-cytosine percentage; TA: Temperature alignment in °C. The superscripts next to each locus indicate the specific cross-amplification. A Haemulon aurolineatum; B Haemulon steindachneri.
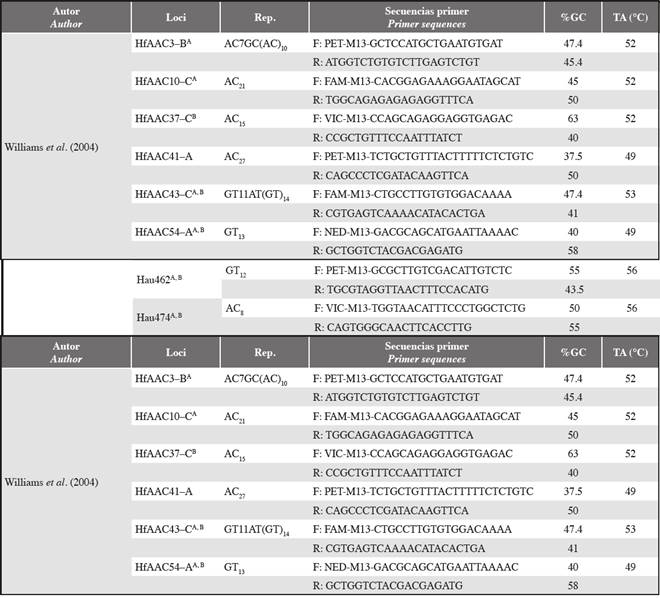
CThe adequate conditions for PCR amplification of the microsatellites were standardized using a ProFlex thermal cycler (ThermoFisher Scientific). The PCR program was based on the conditions required to hybridize the universal M13 primer: one cycle at 94°C for 2 min; 30 cycles of 94°C for 15 s, TA: 48-60°C for 15 s, 72°C for 45 s, 94°C for 15 s; 8 cycles of 53°C for 5 s, 72°C for 45 s; a cycle of 72°C for 15 min and a cycle at 15°C for 15 min. The final concentrations of each reagent were 1X BD buffer, 0.3 mM dNTPs, 1.5 mM MgCl2, 0.25 μM Primer M13 (NED, VIC, PET or FAM), 0.25 μM reverse primer, 0.06 forward primer and 1 U of Taq polymerase. The HfAAC37 and HfAAC3 microsatellites required a mixture of 1X BD buffer, 0.3 mM dNTPs, 2.5 mM MgCl2, 0.16 μM Primer M13 (NED, VIC, PET or FAM), 0.16 μM Primer-R, 0.04 Primer-F and 0.5 U/μl Taq polymerase. The final reaction volume was 10 Verification of the amplifications was performed by electrophoresis in 2.5% agarose gel. The PCR products from these tests were analyzed by capillary electrophoresis in an ABI3130 sequencer, and genotyping was performed with the program Geneious 9.0 (Drummond et al., 2010).
To detect possible genotyping errors (presence of null alleles), the program Micro Checker (Van Oosterhout et al., 2004) was used with 1000 bootstrap simulations at a confidence level of 95%. The deviation from Hardy Weinberg equilibrium (HWE) was estimated by the exact Raymond and Rousset test (1995a) implemented in program GENEPOP v4.2 with 1000 Markov chain Monte Carlo (MCMC) interactions. The linkage disequilibrium was estimated with Fisher's probability test (Raymond and Rousset, 1995b) in GENEPOP v4.2 (Rousset, 2008) with 1000 MCMC interactions using Bonferroni correction.
Of 14 analyzed loci, 10 amplified in both species; it was not possible to amplify the loci HfAAC41-A, Hau034 and Hau075. A high degree of polymorphism was observed: 15 alleles per locus in H. aurolineatum and 21 alleles per locus in H. steindachneri, on average (Figure 1). The HfAAC10-C locus was monomorphic in H. steindachneri. In H. aurolineatum, no loci were found that significantly deviated from HWE, nor were there any null alleles or stuttering in the genotyping (Figure 1b). A significant deviation from HWE was found in H. steindachneri due to a deficit of heterozygotes in the loci Hau220, Hau474 and HfAAC54-A, the latter of which presented null alleles (Fig. 1b). In H. aurolineatum, linkage disequilibrium were detected between the loci pairs Hau315 - HfAAC54-C (p = 0.000), Hau220 - HfAAC3-B (p = 0.000) and HfAAC10-C -Hau474 (p = 0.000), whereas no linkage disequilibrium was found in H. steindachneri (p > 0.05).
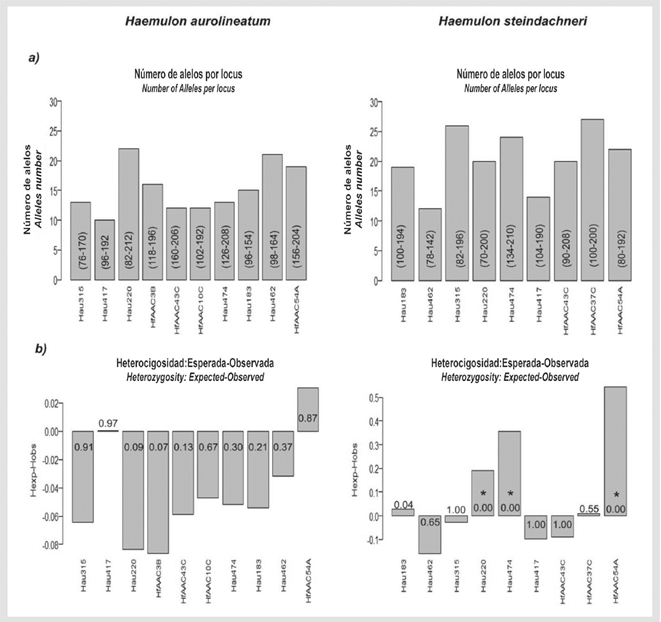
Figure 1 Cross-amplification results for Haemulon aurolineatum and Haemulon steindachneri. a) Number of alleles per locus: for each bar, the size of the alleles in base pairs (bp) is in parentheses. b) Heterozygosity: the p-value is within each bar. *Denotes significant deviation from Hardy Weinberg equilibrium after Bonferroni correction.
This study is the first to test cross-amplification in H. steindachneri, and it provides information about the use of microsatellites in H. aurolineatum. The microsatellites proposed by Williams et al. (2004) were mostly highly polymorphic, but due to the complexity in the type of microsatellite repeats, problems related to null alleles can occur, as in HfAAC54 in H. steindachneri, although this locus did not present null alleles in either H. aurolineatum or H. flavolineatum (Williams et al. 2004).
Therefore, the microsatellites isolated by Williams et al. (2004) and Turner et al. (2005) can successfully be used in population genetics studies of H. aurolineatum and H. steindachneri. Due to their high degree of polymorphism, these microsatellites are high-resolution tools to answer questions of genetic variability in these important ishery species in the Colombian Caribbean.











 texto en
texto en 


