Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Boletín de Investigaciones Marinas y Costeras - INVEMAR
Print version ISSN 0122-9761
Bol. Invest. Mar. Cost. vol.47 no.2 Santa Marta July/Dec. 2018
https://doi.org/10.25268/bimc.invemar.2018.47.2.746
Sección
Bacterial community structure in different tissues of the wild Lobatus gigas (Linnaeus, 1758) from the Caribbean Seaflower Biosphera Reserve
1 Facultad de Ciencias, Universidad Nacional de Colombia, sede Medellín, Colombia. Grupo de Microbiodiversidad y Bioprospección (Microbiop), Universidad Nacional de Colombia-Sede Medellín-Facultad de Ciencias-Escuela de Biociencias-Laboratorio de Biología Celular y Molecular, Calle 59A No 63 - 20 Bloque 19 A Laboratorio 310, Medellín, 050034 Colombia.
The microbial diversity of Lobatus gigas has not been thoroughly studied despite of them is a specie endangered. Knowledge of microbiota may help to improve the conservation and cultivation of this species. The objective of this study was to evaluate the bacterial populations associated with the gonad and the gut compartments of the wild endangered L. gigas from the Caribbean Seaflower Biosphere Reserve, using microbiological methods and culture-independent molecular tools. The genetic profiles of the bacterial populations were generated and Temporal Temperature Gradient Electrophoresis (TTGE) was used to compare them with total DNA. A genetic and statistical analysis of the bacterial communities revealed a low level of diversity in gonad tissue based on the number of bands detected using TTGE. In addition, statistical differences in bacterial community structure were found between the foregut and hindgut tissue of L. gigas. The dominant phylogenetic affiliations of the gonad bacteria, as determined using 16S rRNA gene sequencing, belong to Ralstonia (50%). The possible involvement of this genus in the reproduction and development of the conch is discussed. On the other hand, the bacterial phylotypes from foregut and hindgut included members of Alphaproteobactera (12.5%), Betaproteobacteria (12.5%), Gammaproteobacteria (12.5%), Bacilli (31.25%), Clostridia (6.25%), Actinobacteria (6.25%), Mollicutes (6.25%) and Deinococci (6.25%) classes. Knowing the composition of the gonad and foregut and hindgut bacteria of L. gigas is the first step toward exploring the proper management of this species, as well as provides useful information to future researches that allow a better understanding of the role of these bacterial populations in the health and reproductive rate of L. gigas.
KEYWORDS: Lobatus gigas, 16S rRNA gene; Bacterial Community; Gonad; gut
La diversidad microbiana de Lobatus gigas no se ha estudiado a fondo a pesar de que se trata de una especie en peligro de extinción. El conocimiento de la microbiota puede ayudar a mejorar la conservación y el cultivo de esta especie. El objetivo de este estudio fue evaluar las poblaciones bacterianas asociadas con la gónada y en los compartimentos intestinales de L. gigas en peligro de extinción de la Reserva de la Biosfera Seaflower del Caribe, utilizando métodos microbiológicos y herramientas moleculares independientes del cultivo. Se generaron los perfiles genéticos de las poblaciones bacterianas y se utilizó la Electroforesis Gradual de Gradiente de Temperatura (TTGE) para compararlos con el ADN total. Un análisis genético y estadístico de las comunidades bacterianas reveló un bajo nivel de diversidad en el tejido de las gónadas en función del número de bandas detectadas mediante TTGE. Además, se encontraron diferencias estadísticas en la estructura de la comunidad bacteriana entre el intestino anterior y el tejido del intestino posterior de L. gigas. Las afiliaciones filogenéticas dominantes de las bacterias en la gónada, según se determinó usando la secuenciación del gen RNAr 16S, pertenecen a Ralstonia (50%). Se discute la posible participación de este género en la reproducción y desarrollo del caracol. Por otro lado, los filotipos bacterianos del intestino anterior y del intestino posterior incluyeron miembros de las clases Alphaproteobactera (12.5%), Betaproteobacteria (12.5%), Gammaproteobacteria (12.5%), Bacilli (31.25%), Clostridia (6.25%), Actinobacteria (6.25%), Mollicutes (6.25%) y Deinococci (6.25%). Conocer la composición bacteriana de la gónada y del intestino anterior y posterior de L. gigas es el primer paso para explorar el manejo adecuado de esta especie, y proporciona información útil para futuras investigaciones que permitan una mejor comprensión del papel de estas poblaciones bacterianas en la salud y la tasa reproductiva de L. gigas.
PALABRAS CLAVE: Lobatus gigas; Gen RNAr 16S; Comunidad Bacteriana; Gónada; Intestino
INTRODUCTION
The Queen conch, Lobatus gigas, is a species of ecological and economic importance throughout its distribution area (Catarci, 2004). This species plays an important role in the ecological interactions of the near shore marine populations, as one of the main herbivores of seagrass, epiphyte algae and detritus. Additionally, it is a commensal organism, serves as a food source for a wide variety of marine organisms, and competes with them for resources (Stoner and Waite, 1991; Tewfik, 1997; Catarci, 2004). Furthermore, it is a fishery resource with economic value in the marine food trade. Thus, many countries are required to establish policies to regulate its exploitation by commercializing it in a sustainable way without jeopardizing its conservation. Nonetheless, despite the design and implementation of national and international management measures to control the overexploitation and access to this resource (Prada et al., 2008), their population density continues to decline (Ballesteros et al., 2005; Stoner et al., 2012). This condition hinders the proper maintenance of the population, affecting its self-healing abilities and rendering its commercialization unsustainable (Stoner and Ray-Culp, 2000). As a consequence, it has been included in appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) (Daves and Fields, 2004). In order to overcome this situation and to recover the overexploited populations of L. gigas, several alternatives have been established, such as the creation of marine protected areas (MPA’s) (Appeldoorn, 1994 ; Stoner, 1996; Anon, 1999) which allow the preservation of spawning stock at high densities and maintain a haven for adults with higher reproducibility (Anon, 1999). Another alternative is aquaculture with techniques directed to produce juveniles (Brownell, 1977; Creswell, 1994).
In the Seaflower Biosphere Reserve, several conservation measures have been stablished to regulate L. gigas fishing and to improve understanding of its biology (Castro et al., 2007). More specifically, studies have focused mainly on the population genetics (Landínez- García et al., 2011; Márquez et al., 2012), dynamics, and reproductive biology of this mollusk (Aranda et al., 2001; Aranda et al., 2003a, b; Delgado et al., 2004; Castro et al., 2007). But, these conservation alternatives did not consider studies on microbiota, which is a fundamental aspect of successful culture and restoration of marine organisms. For instance, it has been demonstrated in several species of fish and crustaceans that the microbial community influences various host functions including development, digestion, nutrition, disease resistance and immunity (Merrifield and Ringø, 2014), aspects that are fundamental to the health of any organism. Despite this, to date there is very little known about how the microbiota influence the growth and development of the queen conch (Kirjavainen and Gibson, 1999; Kowalik et al., 2006, Pérez et al., 2014). Recently, others have analyzed the queen conch microbiota through culture-dependent tools, genotypic identification (Acosta et al., 2009, Pérez et al., 2014) and culture-independent approaches (Pérez et al., 2014) using gut and secretion samples from wild conch collected near the Isla del Rosario and the Isla Tortugas in Colombia. These studies demonstrated the presence of Pseudoalteromonas sp., Halomonas sp., Psychrobacter sp., Cobetia sp., Pseudomonas sp., Vibrio sp. and Burkholderia sp. (Acosta et al., 2009; Pérez et al., 2014). Cuartas et al., (2018) using histological analysis, 454 pyrosequencing and a combination of PCR amplification and automated Sanger sequencing indicated that the etiological agent of the muscle protrusions is a parasite belonging to the subclass Digenea and is associated with bacterial and fungi clades. Nevertheless, currently, there are no reports about the molecular characterization of bacterial microbiota associated with the compartments of the gonad and the digestive tract of L. gigas.
The digestive tract is a compound ecosystem that contains a dynamic and complex consortium of microorganisms, which play a key role in the nutrition and health of the host (Bäckhed et al., 2004). The gut microbiota is involved in important processes, such as stimulation of epithelial proliferation, development of nutrient metabolism and innate immune response (Rawls et al., 2004; Sommer and Bäckhed, 2013; Thaiss et al., 2016). Thus, knowledge about the bacteria associated with the different gastrointestinal compartments could be useful to control microbiota as a strategy to improve nutrition, prevent pathogenic infections and to develop new methodologies that contribute to the successful culture of this species. To date, there are no studies aimed at identifying the bacterial communities that inhabit the conch gonad and the potential role on the health and the development of this species. Nonetheless, the presence of bacteria in the gonad has been reported in other mollusks, such as, Argopecten purpuratus where the permanent presence of the genera Vibrio, Acinetobacter, Moraxella, Pseudomonas and Cytophaga in the reproductive organs was found (Chavez and Riquelme, 1994; Riquelme et al., 1995a). In the mollusk Dendrodoris nigra the presence of bacteria in the gonad was determined by means of electron microscopy, but these bacteria were not classified (Klussmann-Kolb and Brodie, 1999). Although the gonad microbiota is poorly understood in marine organisms, previous studies using mammals suggests that the reproductive system microbiome is an important site of bacterial-mammalian symbiosis. These bacteria play important roles, such as, providing protection against pathobiont colonization and producing antimicrobial compounds (Reid et al., 2011; Miller et al., 2017; Kindinger et al., 2017).
The main objective of the present work was to estimate the bacterial community structure associated with gonad, and throughout the different sections of the gut of the wild L. gigas from the Caribbean Seaflower Biosphere Reserve, using culture-dependent and culture- independent analysis. Also, we analyzed microbiota of pool and individuals to study inter-individual variation. To our knowledge, the present work is the first examination of bacterial microbiota of the gonad and throughout the gut compartments of queen conch. This approach enables the detection of dominant bacterial communities that could be of importance in the reproduction and development of the queen conch.
MATERIALS AND METHODS
Sample Collection and Processing
Twenty-two samples of L. gigas (queen conch) juvenile specimens of approximately 350 g each, were collected by fishermen in the Colombian Caribbean Sea, San Andres archipelago (12° -16° N and between 78° - 82°O), these samples were provided by the Gobernación del Archipielago de San Andrés, Providencia y Santa Catalina, through the scientific cooperation agreement #083/2012 (Cuartas et al., 2018). The conchs were transported to the laboratory on dry ice, where the gonad (G), foregut (F), and hindgut (H) of each conch was aseptically extracted under cold conditions. Tissues removed from 15 conchs with similar weight were separatedintofivegroupsandeachgroupwashomogenized and referred to as a pool (P) of each tissue. Pooling samples is a common practice to study the gut microbiota in fish (Hovda et al., 2007; Andlid et al., 1998; Romero and Navarrete, 2006). The tissues corresponding to the seven (7) remaining conchs were processed individually (I), to study inter-individual variation (Romero and Navarrete, 2006). Finally, the samples obtained from tissues (P and I) were immediately macerated with liquid nitrogen and then stored at -80ºC until their processing. Since bacterial communities were obtained from each gut compartment that contained epithelium and digested food, the microbiota analyzed was a combination of both autochthonous (able to colonize the epithelial surface or mucus of the host gut) and allochthonous (transient or associated with digestion) bacteria (Figure 1).
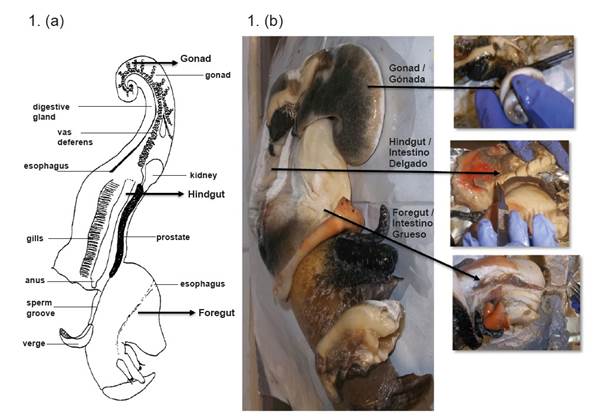
Figure 1 (a). Lobatus gigas anatomy scheme taken from Reed (1996). (b). Dissection and processing of gonad (G), foregut (F) and hindgut of L. gigas.
For the culture-dependent analysis, serial dilutions of homogenates from either P or I conches were plated in marine agar (Difco) and Thiosulfate-citrate-bile salts-sucrose agar-TCBS (Merck), and then the plates were incubated aerobically at 20ºC for 2 days. Colony forming units (CFUs) recovered from the different media were preserved in 20% glycerol at -80ºC and referenced as cultivable fraction (C). The direct fraction (D) refers to the macerated tissue samples frozen at -80ºC without being cultured (G, F, and H). Nomenclature of the samples is summarized in table 1.
Table 1 Nomenclature of samples obtained from the gut compartments and gonad of Lobatus gigas from the biosphere reserve (SEAFLOWER). D= Total DNA from direct fraction, C= DNA from cultivable fraction, I= individual samples, P= pools, G= Gonad, F= foregut and H= hindgut.
| Nomenclatura Nomenclature | Extracción de ADN DNA Extraction | Orígenes Origen | Proceso Process | Muestras Samples |
|---|---|---|---|---|
| CIDI / CFI | Fracción cultivable "C" / Cultivable fraction “C” | Intestino delgado "ID" Foregut “F” | Individual “I” | CIDI2, CIDI3, CIDI4, CIDI6, CIDI16 CFI2, CFI3, CFI4, CFI6, CFI16 |
| CIDP / CFP | Pool "P" | CIDP1, CIDP2, CIDP4, CIDP5 CFP1, CFP2, CFP4, CFP5 | ||
| CIGI / CHI | Intestino grueso "IG" Hindgut “H” | Individual “I” | CIGI2, CIGI3, CIGI6, CIGI16 CHI2, CHI3, CHI6, CHI16 | |
| CIGP / CHP | Pool "P" | CIGP1, CIGP2, CIGP4, CIGP5 CHP1, CHP2, CHP4, CHP5 | ||
| CGI / CGI | Gónada "G" Gonad “G” | Individual “I” | CGI3, CGI4, CGI6, CGI16 | |
| CGP / CGP | Pool "P" | CGP1, CGP2, CGP4, CGP5 | ||
| DIDI / DFI | Fracción directa "D" / Direct fraction “D” | Intestino delgado "ID" Foregut “F” | Individual “I” | DIDI2, DIDI3, DIDI6, DIDI16 DFI2, DFI3, DFI6, DFI16 |
| DIDP / DFP | Pool "P" | DIDP1, DIDP2, DIDP3, DIDP4, DIDP5 DFP1, DFP2, DFP3, DFP4, DFP5 | ||
| DIGI / DHI | Intestino grueso "IG" Hindgut “H” | Individual “I” | DIGI2, DIGI3, DIGI6, DIGI16 DHI2, DHI3, DHI6, DHI16 | |
| DIGP / DHP | Pool "P" | DIGP1, DIGP2, DIGP3, DIGP4, DIGP5 DHP1, DHP2, DHP3, DHP4, DHP5 | ||
| DGI / DGI | Gónada "G" Gonad “G” | Individual “I” | DGI3, DGI4, DGI6, DGI16 | |
| DGP / DGP | Pool "P" | DGP1, DGP2, DGP3, DGP4, DGP5 |
DNA Extraction
The DNA from the cultivable fraction (F) of each sample was extracted with a DNEasy tissue kit (Qiagen, Duesseldford, Germany) using a modified DNEasy DNA extraction protocol to ensure lysis of Gram positive and Gram negative bacteria, as described by Li et al. (2009).
The total genomic DNA from the direct fraction of each tissue was extracted using an Ultraclean soil DNA extraction kit with a prior tissue lysis step, as described by Ó Cuív et al. (2011). Briefly, each dissected tissue from L. gigas was obtained as described above and 300 mg were homogenized with 800 μl of lysis buffer (6 mM Tris-HCl [pH 8], 100 mM EDTA, 1M NaCl) and incubated at 75ºC for 10 minutes to inactivate nucleases. 20 μl of lysozyme were added (200 mg/ml) and incubated overnight at 37ºC. Subsequently, a digestion with 20 μl of proteinase K (20 mg/ml) at 56ºC was performed until complete lysis of each tissue.
PCR amplification
In order to obtain molecular fingerprinting of the bacterial community from the foregut, hindgut and gonad cultured on marine agar (C), the 16S rRNA gene between V3 and V6 region was amplified with specific primers 341F with an extra GC tail (5 ′GCCTACGGGAGGCAGCAG 3′) and 907R (5′CCGTCAATTCMTTTGAGTTT 3′) (McCracken et al., 2001). The PCR was carried out using the conditions described by García et al. (2016).
Amplification of the hypervariable region between V3 and V6 regions of 16S rRNA from the D was performed with a nested PCR. The first PCR was carried out to amplify nearly full-length 16S rRNA gene sequences with universal primers 27F (5`AGAGTTTGATCMTGGCTCAG 3`) and 1492R (5` GTTACCTTGTTACGACTT 3`) following the conditions given by Espejo and Romero (1997). Then, 2.5 μl from the product of the first PCR were used as template DNA in the second reaction, which was performed with the specific primers 341-GC F and 907R (as described previously for cultivable samples) and under the conditions reported by Espejo et al. (1998).
Temporal Temperature Gradient Electrophoresis (TTGE) analysis
The structure of bacterial communities associated with the foregut, hindgut and gonad was revealed using the TTGE technique, which allowed separating fragments of the 16S rRNA gene of approximately 585 bp, with the assumption that each band observed in the banding patterns represents a bacterial species.
The PCR products obtained from both fractions, using the 341F-GC and 907R primers, were separated by TTGE using the DCode Universal Mutation Detection System (BioRad, USA) on 7% (w/v) polyacrylamide, 7M Urea gels in 1.25X TAE buffer for 15h at 55V. The initial and final temperatures were 66ºC and 69º respectively, with a temperature ramp of 0.1ºC per hour. The gels were stained with AgNO3 (AMRESCO, OH, USA) (Sanguinetti et al., 1994). The band patterns obtained for each sample studied were analyzed using GelCompar II (Applied Maths TX, USA) (Rademaker and De Bruijn, 2008, Garcia et al., 2016). The reference lanes from each sample were aligned according to a 100 bp ladder, and a cluster analysis was performed using DICE coefficient (Nei and Li, 1979) and the Unweighted Pair Group Method with Arithmetic Average (UPGMA) (Mohammadi and Prasanna, 2003).
Bacterial community analysis
GelCompar II software (Applied Biosystems, Belgium) was used to analyze TTGE profile patterns from the DNA samples. For this analysis, the presence and absence matrix was used to generate an analysis of similarity (ANOSIM) based on the Bray-Curtis index using PAST software version 2.0 (Hammer et al., 2001). Significant differences in bacterial diversity were assessed with the “Diversity t-test” option of the PAST program. For this analysis, the band number was considered to represent the species number or Taxa (S) and the band intensity was considered to represent the relative abundance or number of individuals of each bacterial species. The “Principal Component Analysis” option was used to view overall similarities and dissimilarities among sampling of the gonad, foregut and hindgut and between the cultivable (C) and direct (D) fraction from each tissue (Chaiyapechara et al., 2012; Pérez et al., 2014, Boon, 2002). This clustering distribution was confirmed by the analysis carried out with the two gel replicates. Finally, the Shannon index (H`) which reflects the diversity of the bacterial community of F, H and G was calculated following the formula: H` = - SPi ln Pi, where Pi is defined as (ni ⁄N), ni is each band present, and N is the sum of all bands (Sun et al., 2011).
TTGE bands with unique migration patterns according to the GenRulerTM 100bp DNA ladder (Thermo Fisher Scientific) and bands with higher intensity in each TTGE, were cleaved and DNA was eluted following the Soak and Crush method described by Sambrook and Russell (2002). Then 5μl from the final elution were taken and re-amplified with primers 341F without GC tail and 907R. Re-amplified products from the C fraction was verified by gel electrophoresis on a 1% agarose gel. The re-amplified products from D were purified using the QIAquick PCR purification kit (QIAGEN CA, U.S.A) and cloned into E. coli DH5α competent cells using a Clone JET TM kit (Thermo Scientific, U.S.A). Ligation was performed following the manufacturer’s indications and transformation was completed as described by Sambrook and Russell (2002). The insert was amplified following the QIAquick purification kit (QIAGEN CA, U.S.A) recommendations with pJET1.1 and pJET 1.2 primers. Amplicons from D and C were sequenced in on an ABI PrimR 3100 Genetic Analyser (Applied Biosystems, CA, USA).
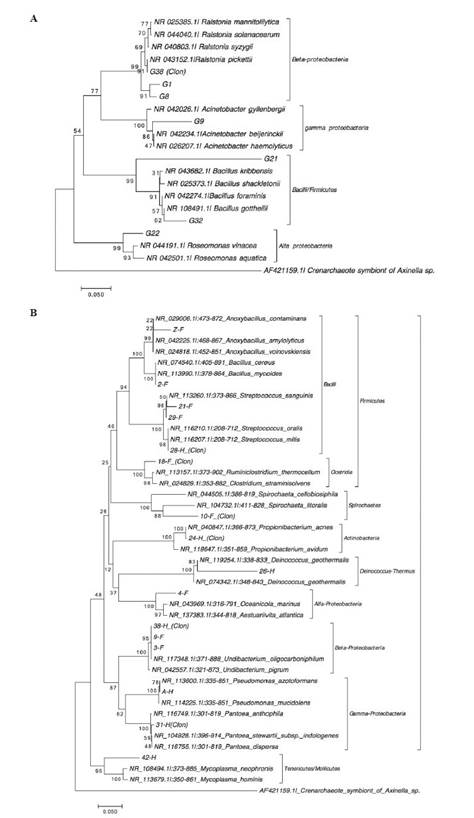
Sequences of 16S rDNA obtained from both fractions were edited using BioEdit ® (Hall, 1999) and the presence of chimeras was assessed with the online software DECIPHER v2.0 described by Wright et al. (2012). The edited sequences were then compared with reference sequences from The National Center for Biotechnology information (NCBI) and Ribosomal Database Project (RDP). To determine the phylogenetic affiliation of the microbial isolates and the excised bands, similarity searches and phylogenetic analysis were performed using the BLASTN and MEGA 7 software (Tamura et al., 2007). Phylogenetic analysis was performed using the Neighbor-Joining method (Saitou and Nei, 1987) with 1000 bootstrap replicates. It was rooted with a 16S rRNA gene sequence from Crenarchaeote symbiont of Axinella sp., in order to improve the topology. The sequences obtained in this study were deposited in GenBank under accession numbers KX891431-KX891450 and KX886796-KAX886797.
RESULTS
Bacterial count
The average cultivable bacterial abundance of gut tract compartments and gonad in each medium is observed in Table 2 and expressed as CFUs per gram of tissue. The averages of bacterial abundance recovered from the gonad and cultured in marine and TCBS agar were 1,21x107 y 2,83x 107 CFUs per gram of tissue, respectively. The TCBS agar counts indicated a similar abundance in both gut compartments (1.01 x 107CFUs per gram of tissue), while on the other hand, the counts from the foregut bacteria cultured in marine agar revealed a concentration of heterotrophic bacteria five times greater than the hindgut.
Table 2 Average bacterial abundance (CFUs/g) from gonad, foregut and hindgut cultured in marine and TCBS agar.
| Tejido Tissue | Agar marino (UFC/g) Marine Agar (CFU/g) | Agar TCBS (UFC/g) Agar TCBS (CFU/g)T |
|---|---|---|
| Intestino delgado Foregut | 5.57*107 | 1.01*107 |
| Intestino grueso Hindgut | 1.10*107 | 1.00*107 |
| Gónada Gonad | 1.21*107 | 2.83*107 |
Analysis of PCR-TTGE profiles
The estimation of the bacterial community that were associated with gonad and gut tissues and between cultivable “C” and direct “D” fraction were evaluated through TTGE profiling of partial 16S rRNAgenes (Figure 2A and Figure 2B). The results of the banding profiles, evaluated using the program GelCompar II, revealed the presence of several migration patterns, these profiles revealed differences between the bacterial communities associated with each tissue and the “D” fraction samples exhibited more complex patterns and more diverse bands compared with those of the cultivable “C” fraction.
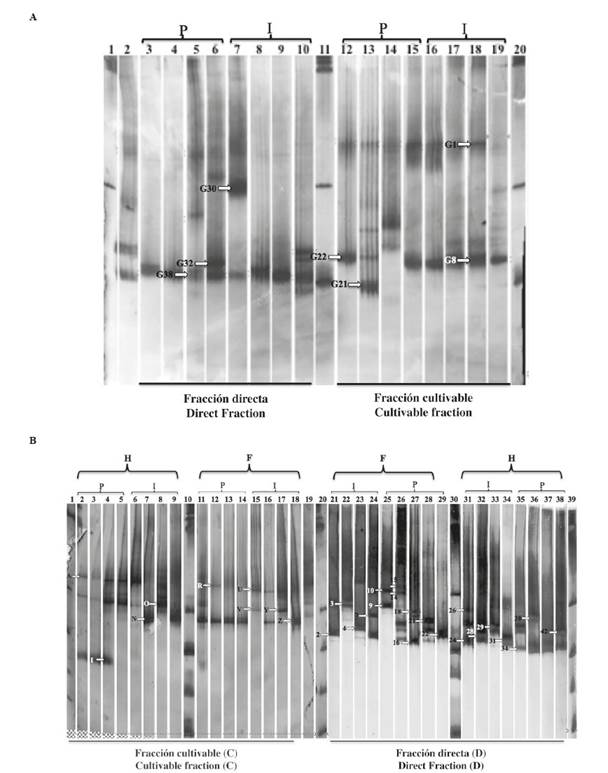
Figure 2 TTGE profile of V3-V6 region of 16S rRNA gene from cultivable and direct fraction gonad (A) and foregut, hindgut (B). The labels above the image indicate the samples origin. “G” for gonad, “F” for foregut and “H” for hindgut, “P” for pool and “I” for individual. The lanes labeled with “L” correspond to markers of 100 bp used as references. The codes above each lane indicate the bands selected for sequencing.
From this gel analysis (Figure 2), 105 (69 of gut and 36 gonad) unique and common DNA bands were selected based on the band intensity. Referring to the gonad of Lobatus gigas, the G38 band showed a predominant population associated with individual and pooled samples from the direct fraction. The cultivable fraction was characterized by the presence of bands G1 and G8 in three pools and four individual samples (Figure 2A). The results of the DNA bands sequences are provided in Table 4.
The genetic profiles and the clustering analysis performed from direct and cultivable fraction of the gonad revealed two different groups according to the fraction, with 22.2% similarity for group A and 27.9% for group B (Figure 3A). Two subgroups were formed in group A from cultivable fraction samples, one of them with a higher than 60% similarity and the other with a similarity level of 50%. Group B contained the samples from the direct fractions (Figure 3A).
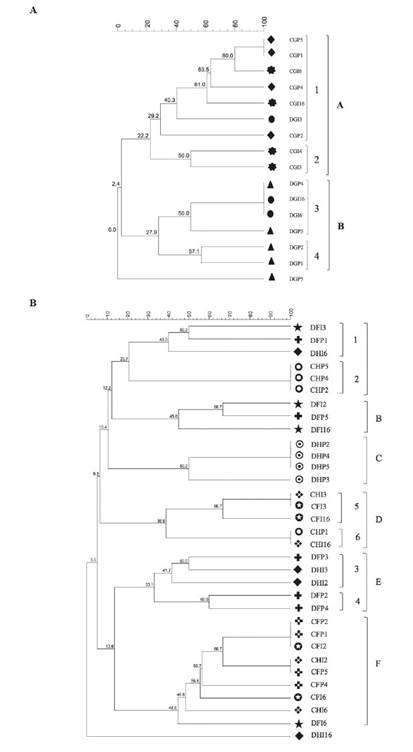
Figure 3 TTGE banding patterns cluster analysis from cultivable “C” and direct “D” fraction of individual “I” and pools “P” samples of gonad “G” (A) and foregut “F” and hindgut “H” (B) of L. gigas. The cluster analysis was performed with Gel compare II ® software using Dice coefficient and Unweighted - Pair Group Method with Arithmetic Mean (UPGMA) (Mohammadi and Prasanna, 2003). The letters from A to F and the numbers represent the groups and the sub-groups generated, respectively.
On the other hand, the genetic profiles with the UPGMA method for the clusters analysis of foregut and hindgut showed six groups A, B, C, D, E and F with similarity percentages of 20.7, 45, 38.9, 33.1 and 44.6 %, respectively (Figure 3B). Group A is comprised of hindgut and foregut samples from both cultivable and direct fraction, however, two subgroups were also formed according to the fraction where the samples came from (Figure 3B, subgroup 1 and 2). Group B clustered two individual samples (DFI2 and DFI16) and one pool (DFP5) from the direct fraction of the foregut with a similarity percentage of 45%. Group C contained all samples from the direct fraction of the hindgut, with a similarity percentage higher than 50%. In cluster D the samples from cultivable fraction were grouped with similarity percentages of 38.9% and 44.6% respectively, and a high similarity percentage was observed in the bacterial population from the foregut. Furthermore, the samples from direct fractions were clustered mainly in the groups B, C and E, with a similarity of 45, 50 and 33.1%, respectively. Group E contained hindgut and foregut samples from direct fractions, with similarity levels higher than 33.1%.
Variations in bacterial communities on different tissue (G, F, and H) observed in the UPGMA analyses, were verified through a principal component analyses PCA (Figure 4).
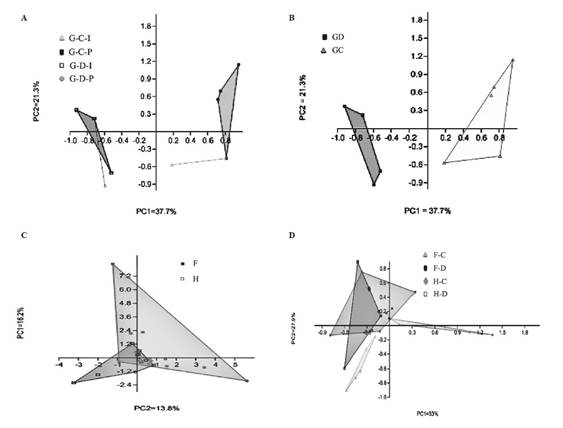
Figure 4 (A) Principal Components Analysis (PCA) of individual samples and pools of the gonad “G” obtained from “C” cultivable and “D” direct fraction. (B) PCA of “G” gonad samples obtained from “C” cultivable and “D” direct fraction. (C) PCA to evaluate the differences between bacterial communities associated with “F” foregut and “H” hindgut of Lobatus gigas. (D). PCA of gut compartments obtained from “C” cultivable and “D” direct fraction.
According to the binary matrix of data on the presence (1) and absence (0) of bands, 36 bands were detected on the gels of the gonad. A band-based binary presence/absence matrix was calculated by applying the Dice similarity coefficient and used for an analysis of similarity (ANOSIM) based on the Bray-Curtis coefficient, which enables significance testing of the data groups (Hammer et al., 2001). Significant differences were found between the samples associated with “C” cultivable and “D” direct fraction [R = 0.868 (Bray Curtis) and (Jaccard), (P < 0.004 in both analyses)]. Moreover, differences between bacterial community compositions of the cultivable fraction of individual samples and pools were demonstrated (Figure 4A). The Shannon Index (H`) analysis showed a difference between the bacterial communities associated with “C” cultivable and “D” direct fraction (Table 3). Also, the analysis showed that bacterial populations associated with individual gonad tissue samples are less related than those obtained from pools of gonad tissue and that this is true for both fractions.
Table 3 Shannon index (H`) calculated from the TTGE profiles of V3-V6 region of 16S rRNA for each tissue, each fraction.
| Índice de Shannon (H`) Shannon Index (H`) | |||
|---|---|---|---|
| Tejidos Tissues | Fracción C C Fraction | Fracción D D Fraction | Fracciones C y D C and D Fractions |
| Intestino delgado (ID) / Foregut (F) | 1.4 | 2.5 | 2.5 |
| Intestino grueso (IG) / Hindgut (H) | 1.7 | 1.7 | 2.4 |
| ID e IG / F and H | 2.2 | 2.7 | 2.9 |
| Gónada / Gonad | 1.6 | 1.9 | 2.3 |
Furthermore, the results of the analysis shownthat 69 bands were detected on the gels of the gut tissue. PCA observed in figure 4C and the analysis of similarity (ANOSIM), showed significant differences in the bacterial community associated with the foregut (F) and hindgut (H) [R=0.1406 and (Bray Curtis) and (Jaccard), (P < 0.0018 in both analyses)]. The differences among bacterial communities related to cultivable and direct fraction from each compartment is observed in the PCA (Figure 4D). Significant differences were found between the samples associated with “C” cultivable and “D” direct fraction of the foregut (F) [R = 0.3479 (Jaccard) and R= 0.3479 (Bray-Curtis), (P value < 0.0024 in both analyses)], and hindgut (H)[R = 0.4643 (Jaccard) and R= 0.4643 (Bray Curtis), (P value < 0.0003) ].
The H` values calculated from G, F, and H (taking into account C and D fraction) revealed that the foregut has the most diverse bacterial community while the gonad has the least diverse. When H` was calculated for each tissue and for C and D separately, it was possible observe that the diversity is higher in D fraction than the C fraction, except for hindgut tissue which kept its diversity for both approaches (Table 3).
Sequencing and phylogenetic analysis of the bands (≈400 bp) excised from TTGE gel revealed several different phylogenetic groups for each sampling tissue (gonad, foregut and hindgut) according to their similarity with 16S rRNA gene sequences held in public databases (Table 4, Figure 5). These belonged to Alphaproteobacteria, Gammaproteobacteria and Bacilli (Table 4, Figure 5A) for the gonad, and the most common species was Ralstonia pikettii, which was observed in both cultivable and direct fraction (Figure 4A). In the gut compartments, the results of partial sequences of the 16S rRNA gene revealed high similarity with bacteria belonging to Alphaproteobacteria (12.5%), Betaproteobacteria (12.5%), Gammaproteobacteria (12.5%), Bacilli (31.25%), Clostridia (6.25%), Actinobacteria (6.25%), Mollicutes (6.25%) and Deinococci (6.25%) and unclassified bacteria (Table 4, Figure 5B).
Table 4 Taxonomic affiliations of DNA partial sequences obtained from TTGE bands of L. gigas gonad, foregut, and hindgut.
| Origen Origin | Banda TTGE TTGE Band | Nº de acceso Accession N° | Porcentaje de similitud (BlastN) Percent Similarity (BlastN) | Filo Affiliation Phylum | Clase Class | Secuencia más cercana Closest sequence | |
|---|---|---|---|---|---|---|---|
| DIDI6 / DFI6 | 4F | KX891442 | 93% | α-Proteobacteria | Oceanicola marinus. NR_043969.1 | ||
| DIDP1 / DFP1 | 9F | KX891443 | 98% | β-Proteobacteria | Undibacterium oligocarboniphilum. NR_117348.1 | ||
| DIDI3 / DFI3 | 3F | KX891440 | 97% | Proteobacteria | |||
| DIGP3 / DPH3 | Clon 38H | KX891439 | 98% | ||||
| CIGP5 / CHP5 | AH | KX891445 | 99% | γ-Proteobacteria | Pseudomonas azotoformans. NR_113600.1 | ||
| DIGI16 / DHI16 | Clon 31H | KX891438 | 99% | Pantoea stewartii. NR_104928.1 | |||
| DIDP3 / DFP3 | Clon 18F | KX891432 | 95% | Firmicutes | Clostridia | Clostridium straminisolvens. NR_024829.1 | |
| DIDP4 / DFP4 | 21F | KX891442 | 100% | Bacilli | Streptococcus sanguinis. NR_113260.1 | ||
| DIGI6 / DHI6 | 29H | KX891431 | 100% | Streptococcus sanguinis. NR_074974.1 | |||
| DIGI3 / DHI3 | Clon 28H | KX891436 | 100% | Streptococcus mitis. NR_116207.1 | |||
| CIDI2 / CFI2 | ZF | KX891444 | 96% | Anoxibacillus amylolyticus. NR_042225.1 | |||
| DIDI2 / DFI2 | 2F | KX891437 | 100% | Bacillus cereus. NR_074540.1 | |||
| DIGI2 / DHI2 | Clon 24H | KX891434 | 99% | Actinobacteria | Actinobacteria | Propionibacterium acnes. NR_040847.1 | |
| 26H | KX891435 | 88% | Deinococcus- Thermus | Deinococci | Deinococcus geothermalis. NR_074342.1 | ||
| DIGP5 / DHP5 | 42H | KX891441 | 88% | Tenericutes | Mollicutes | Mycoplasma neophronis. NR_108494.1 | |
| DIDP1 / DFP1 | Clon 10F | Secuencia de la quimera | 93% | Spirochaetae | Spirochaetia | Spirochaeta litoralis. NR_104732.1 | |
| CGI4 / CGI4 | G1 | KX891446 | 96% | Ralstonia pickettii. NR_043152.1 | |||
| 96% | Ralstonia pickettii. NR_102967.1 | ||||||
| G8 | KX891447 | 96% | β-Proteobacteria | Ralstonia pickettii. NR_043152.1 | |||
| 96% | Ralstonia mannitolilytica. NR_025385.1 | ||||||
| DGP5 / DGP5 | Clon G38 | KX886796 | 99% | Proteobacteria | Ralstonia pickettii. NR_043152.1 | ||
| 98% | Ralstonia mannitolilytica. NR_025385.1 | ||||||
| CGP5 / CGP5 | G22 | KX891450 | 95% | α-Proteobacteria | Roseomonas aquatica. NR_042501.1 | ||
| 94% | Roseomonas vinacea. NR_044191.1 | ||||||
| CGP4 / CGP4 | G21 | KX886797 | 83% | Firmicutes | Bacilli | Bacillus gotheilli. NR_108491.1 | |
| 83% | Bacillus foraminis. NR_042274.1 | ||||||
| DGP2 / DGP2 | G32 | KX891449 | 94% | Bacillus lichenoformis. NR_074923.1 | |||
| 94% | Bacillus shackletonii. NR_025373.1 | ||||||
DISCUSSION
L. gigas microbiota from the Colombian Caribbean has been characterized by recent studies, with the goal of study the bacterial diversity associated with both wild and captive conchs (Pérez et al., 2014; Acosta et al., 2009; Rodriguez et al., 2011). In this study we conduct one of the first exploratory investigations into consideration the different tissues to observe the dominant bacterial community in of L. gigas. We analyzed the bacterial composition of the gonad and the gut (foregut and hindgut) of the wild queen conch from the Seaflower biosphere reserve by using different microbiological and molecular methods. When we compared the cultured bacteria and a molecular approach based on analysis of DNA extract direct from the sample seems to be a complementary strategy to determine the principal components in the microbial community of queen conch.
The analysis performed using conventional culture techniques, such as counting CFUs in marine and TCBS agar, allowed us to establish the presence of heterotrophic bacteria and bacteria belonging to the family Vibrionacea in the gonad and in the gut compartments of the queen conch. The TCBS agar count suggests an abundance of vibrios with a total of 2.83 x 107 CFUs per gram in gonad tissue and 1.01 x 107 CFUs per gram of tissue for the gut compartments analyzed; nevertheless, to date no studies have described the gonad microbiota of the conch. Previously, Avendaño-Herrera et al. (2001) reported a load of heterotrophic bacteria in the bivalve Argopecten purpuratus gonad in captivity; they counted 4 x 104 CFUs per gram and a Vibrionacea count of 3 x 102 CFUs per gram. Our results contrast with those describe by Avendaño-Herrera et al. (2001) and could be due to the captivity conditions under which the A. purpuratus studies were performed. The affinity of vibrios to the gonad tissue has been previously described in the mollusk Crassostrea gigas and it has been related to episodes of mortality of this species (De Decker et al., 2011). Studies aimed at establishing the identity of vibrios associated with the gonad, foregut and hindgut and its possible effects on mollusk development are necessary. Also, the bacteria belonging to the family Enterobacteriaceae and the genera Aeromonas and Pseudomonas can develop in the TCBS medium, and they could interfere with bacterial counts, generating an overestimation in the total vibrio abundance present in the tissue analyzed.
The analysis of TTGE profiles showed similar bacterial communities among the individual and pool samples in the gonad tissue (Figure 2A), with dominance of two or three bands per profile. These results could be related with the autochthonous bacteria of this mollusk tissue. The differences in the banding profiles observed between individual and pooled samples from gonad, foregut and hindgut were verified with a PCA. The analysis highly suggests that there is a greater difference in bacterial populations associated with individual samples than those associated with pooled samples from both fractions (C and D) (Figure 3). Similar results have been reported in different studies with marine organisms. In these studies individual variation was observed even among fish raised under similar environmental conditions, with similar genetic background, and fed the same diet, emphasizing the influence of the host on bacterial diversity (Spor et al., 2011; McKnite et al., 2012; Navarrete et al., 2012). Besides this, banding patterns from pools were similar in some samples, suggesting the presence of a dominant bacterial population in these tissues. However, these results could lead to biased conclusions, due to the possibility that the dominant bacteria present in one individual could be interpreted as common bacteria for all the individuals analyzed in the sample (Reveco et al., 2014).
Meanwhile, the results revealed the presence of a more complex and diverse community associated with the conch foregut when compared to the hindgut. Important shifts in the bacterial community structure were also observed between cultivable fraction (C) and direct fraction (D). Significant differences between both gut compartments were found with an average percentage of dissimilarity of 94.41%. These results were verified with the principal components analysis presented in figure 4C, which suggests that the foregut and hindgut have been colonized by different bacterial communities, this fact was supported with diversity index (H`), which showed a higher diversity in foregut than in hindgut (Table 3). These differences could be associated with metabolic processes carried out specifically in each compartment. Studies on fish suggest that variation in microbial diversity among gut compartments could be due to differences in pH and protease activity in the lumen (Yu et al., 2007; Yúfera and Darías, 2007). Furthermore, differences in the pH of the Epinephelus coioides fish stomach, pyloric caeca and intestine have been reported (Yu et al., 2007). These physicochemical parameters might be associated with the changes observed in the microbiota of different gut compartments, which could be related to specific functions in each gut and the ecological conditions of the habitat where L. gigas feeds.
According to the phylogenetic analysis, sequences from the gonad, indicated that the bacterial community has been colonized mainly by bacteria belonging to the genus Ralstonia, which were represented by several TTGE bands in the direct and cultivable fraction of pooled and individual samples (Figure 2A, Table 4). This finding suggests that the gonad could be a site of bacteria symbiosis in the queen conch. Although little is known about gonad microbiota in marine organisms and its interaction with the host, previous studies of the microbiome in mammals have demonstrated that in the reproductive system there is a mammal-bacteria symbiosis and this interaction may help defend the host against infectious diseases and improve its reproductive health (Miller et al., 2017; Kindinger et al., 2017).
In the case of the foregut and hindgut, phylogenetic analysis revealed differences between the two compartments (Table 4 and Figure 5B). The species found in both compartments belong to the phylum Firmicutes and Actinobacteria, while the phylum Proteobacteria, Deinococcus - Thermus, and Tenericutes were only found in the conch hindgut. These results are consistent with those reported in gut samples of the queen conch from Islas del Rosario (Acosta et al., 2009; Pérez et al., 2014) and other marine organisms (Sun et al., 2011; Trabal et al., 2012; Chaiyapechara et al., 2012). Although there are differences in the methodologies employed, the results are related to a high bacterial diversity associated with this mollusk.
Bacteria of the genus Bacillus and Propionibacterium have been characterized as probiotics in fish and marine crustaceans (Nakagawa et al., 2007; Lan et al et al., 2007; Cousin et al., 2010). More specifically, the genus Bacillus has been established as a biological agent against Aeromonas hydrophila (Lalloo et al., 2007, 2008) and the members of this genus produce a wide range of antagonistic compounds that have been found in the gut tract of crustaceans and fish (Balcázar et al., 2006). The presence of Pseudomonas in the foregut of the queen conch coincides with the results reported in other mollusks and in different marine organisms such as crustaceans and fish (Sfanos et al., 2005; Romero and Navarrete, 2006; Trabal et al., 2012). It is important to notice that members of this genus isolated from marine organisms have been recognized for their antibiotic activity against a broad spectrum of microorganisms (Gram, 1993; Jayatilake et al., 1996; Zheng et al., 2005). These results suggest that the presence of Pseudomonas libanensis could be related to beneficial effects and it could contribute to the survival of L. gigas by defending it against potential pathogens that may affect its health and development. Meanwhile, the genus Streptococcus (Table 4 and Figure 5B) has been associated with several diseases in fish and crustaceans, and it has been identified as the causative agent of the slow development disease in the crustacean Carcinus mediterraneus (Pappalardo and Boemare, 1982). The genus Undibacterium and Oceanicola found in the foregut and hindgut of the queen conch (Table 4) has also been recently reported in the gut of wild crustaceans Penaeus monodon samples (Rungrassamee et al., 2014). It is remarkable that Oceanicola marinus is a common specie in marine environments (Lin et al., 2007; Chunyan et al., 2012).
Differences between the results obtained from cultivable and direct fraction analysis were observed in the conch gonad, foregut and hindgut. It is possible that these differences are due to the culture dependent techniques which favor the growth of bacteria able to grow on synthetic media, regardless whether these bacteria are dominant in the sample. It has been shown that these bacteria represent only 1% of the total population, generating an underestimation of the actual microbial population diversity (Hansen and Olafsen, 1999; Suau et al., 1999). Further studies based on culture-independent methods have demonstrated a less biased picture of the bacterial population present in environmental samples than culture-dependent methods (Amann et al., 1995). Nonetheless, it is necessary to take into account that techniques based on ribosomal genes may be affected by low concentrations of DNA from different species or the presence of high concentrations of DNA that are competing during PCR reactions (Ogier et al., 2002), which can affect the detection of some species in the community (Kisand and Wikner, 2003). That is why; it is necessary to implement both approaches (culture-dependent and culture-independent) in the study of microbiota from natural samples, in order to recover as much information as possible about the bacterial species present in the samples analyzed (Kisand and Wikner, 2003).
This research established the bacterial community associated with gonad and foregut and hindgut of L. gigas. Regarding the gonad, the results suggest that this organ has a stable microbiota that is mainly dominated by members of the genus Ralstonia that could be endosymbionts of this tissue. Additionally, the presence of vibrios should be verified and it is necessary to determine their effect on the gonad. But, more comprehensive studies are required to determine the role played by this bacterial community in the health and the reproductive cycle of the L. gigas. In the case of the foregut and hindgut, phylogenetic analysis revealed differences between the two compartments. However, to establish the effect of these communities on the different compartments of the conch, further investigations are necessary. Overall, the findings reported here could be used for the development of strategies to increase nutrition, prevent pathogens and contribute to the conservation of this vulnerable species from the Seaflower Biosphere Reserve in the Colombian Caribbean.
ACKNOWLEDGMENTS
We thank to the Universidad Nacional de Colombia and Secretaría de Agricultura y Pesca del Archipiélago de San Andrés, Providencia y Santa Catalina, Isla de San Andrés, through the projects 20101009144 and 201010011106, for their financial support. According to the Colombian legislative framework of the Environmental Ministry of Colombia, this project is subscribed to “Permiso Marco No. 0255” granted by the Autoridad Nacional de Licencias Ambientales (ANLA) to the Universidad Nacional de Colombia on March 14, 2014. Special thanks to Erick Castro, Jaisón Cuartas and Luz Pineda for all technical support provided and PhD Gloria Cadavid for her unconditional support.
BIBLIOGRAFÍA
Acosta, E.A., E. Gómez, M. Romero-Tabarez, G.E. Cadavid-Restrepo and C.X. Moreno-Herrera. 2009. Molecular identification of bacterial populations associated to queen conch (Strombus gigas)from Colombian Caribbean. Acta Biol. Col., 14: 83-96. [ Links ]
Amann, R.I., W. Ludwig and K.H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev., 59: 143-169. [ Links ]
Andlid, T., R. Vazquez-Juarez and L. Gustafsson. 1998. Yeasts isolated from the intestine of Rainbow trout adhere to and grow in intestinal mucus. Mol. Mar. Biol. Biotechnol., 7: 115-126. [ Links ]
Anonymous. 1999. Report on the /Queen Conch/ Stock Assessment and Management Workshop. Belize City, Belize, 15-22. Caribbean Fisheries Management Council and CARICOM Fisheries Resources Assessment and Management Programme. http://caribbeanfmc.com/reports-sci-docs/BelizeConchWGReportFinal.PDF. 20/06/2017. [ Links ]
Appeldoorn, R.S. and B.Q. Rodríguez. 1994. Queen conch biology, fisheries and mariculture. Fundación Científica. Los Roques, Caracas, Venezuela. 301-319. [ Links ]
Aranda D., C.E. Baqueiro, I. Martínez, R.I. Ochoa and T. Brulé. 2001. Reproductive patterns of Strombus gigas from Alacranes reef versus Chinchorro bank of Mexico. Gulf Caribbean Fisheries Institute, 54: 202-224. [ Links ]
Aranda, D.A., C.E. Baqueiro , I. Martínez , R.I. Ochoa and T. Brulé . 2003a. Gonad behavior during peak reproduction period of Strombus gigas from Banco Chinchorro. Bull. Mar. Sci., 73: 241-248. [ Links ]
Aranda, D.A ., C.E. Baqueiro , I. Martínez , R.I. Ochoa and T. Brulé . 2003b. A review of the reproductive patterns of gastropod mollusks from Mexico. Bull. Mar. Sci ., 73: 629 - 641. [ Links ]
Avendaño-Herrera, R.E., M. Dekovic y C.E. Riquelme. 2001. Establecimiento de bacterias benéficas en el tracto digestivo y gónada de adultos de Argopecten purpuratus (Lamarck 1819) en cultivo masivo. Rev. Biol. Mar Ocean., 36: 31-41. [ Links ]
Bäckhed, F., H. Ding, T. Wang, L.V. Hooper, G.Y. Koh, A. Nagy, C.F. Semenkovich and J.I. Gordon. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A., 101: 15718-15723. [ Links ]
Balcázar, L., O. Decamp, D. Vendrell, I. De Blas and I. Ruiz-Zarzuela. 2006. Health and nutritional properties of probiotics in fish and shellfish. Microb. Ecol. Health Dis., 18: 65-70. doi:10.1080/08910600600799497. [ Links ]
Ballesteros, F., C. García, M. Rueda, K. Gómez and Mejia. 2005. Relative. Abundance and Fishery Characterization of Queen Conch Strombus gigas (Mesogastropoda- Strombidae) in the Archipielago of San Bernardo, Colombian Caribbean. Proc. Gulf Car. Fish. Inst., 58: 393-398. [ Links ]
Boon, N. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol., 39: 101-112. http://dx.doi.org/10.1016/s0168-6496(01)00198- 2. [ Links ]
Brownell, W.N. 1977. Reproduction, laboratory culture and growth of Strombus gigas, Strombus costatus and Strombus pugilis in Los Roques, Venezuela. Bull. Mar. Sci ., 27: 668-680. [ Links ]
Castro, E., H. Bent, C. Ballesteros and M. Prada. 2007. Large pelagics in the southern section of the seaflower marine protected area, San Andres archipelago, Colombia: a fishery in expansion. Gulf Car. Res., 19: 131-139. [ Links ]
Catarci, C. 2004. World markets and industry of selected commercially-exploited aquatic species with an international conservation profile. FAO Fisheries Circular. No. 990. Rome, FAO. [ Links ]
Chaiyapechara, S., W. Rungrassamee, I. Suriyachay, Y. Kuncharin, A. Klanchui, N. Karoonuthaisiri and P. Jiravanichpaisal. 2012. Bacterial Community Associated with the Intestinal Tract of P. monodon in Commercial Farms. Microb. Ecol., 63: 938-953. doi:10.1007/s00248-011-9936-2. [ Links ]
Chavez, P. y C. Riquelme. 1994. Análisis de la calidad bacteriológica en reproductores de Argopecten purpuratus (Lamarck, 1819) para su uso en acuicultura. Rev. Lat. Acuicult., 43:96-99. [ Links ]
Chunyan, X.U., Y.A.N. Qingpi and M.A. Ying. 2012. Microorganisms colonizing surface in coastal marine water as revealed by 16S rRNA gene clone library analysis. Afr. J. Microbiol. Res., 6: 7271-7277. [ Links ]
Cousin, F.J., D.D.G. Mater, B. Foligne and G. Jan. 2010. Dairy propionibacteria as human probiotics: A review of recent evidence. Dairy Science and Technology, 91: 1-26. doi:10.1051/dst/2010032. [ Links ]
Creswell, L. 1994. An historical overview of queen conch mariculture: 223-230. Appeldoorn, R.S . and B. Rodríguez. (Eds.)., Queen Conch Biology, Fisheries and Mariculture. Fund. Cient. Los Roques, Caracas. Venezuela. 223-230. [ Links ]
Cuartas, J., J. Alzate, C.X. Moreno-Herrera and E. Marquez. 2018. Metagenomic of orange colored protrusions from the muscle of Queen Conh Lobatus gigas (Linnaeus, 1758) PeerJ, 6:e4307. https://doi.org/10.7717/peerj.4307. [ Links ]
Daves, N. and J. Fields. 2004. Recent developments in CITES concerning the international trade in queen conch (Strombus gigas). Proc. Gulf. Caribb. Fish. Inst., 763-770. [ Links ]
De Decker, S., J. Normand, D. Saulnier, F. Pernet, S. Castagnet and P. Boudry. 2011. Responses of diploid and triploid Pacific oysters Crassostrea gigas to Vibrio infection in relation to their reproductive status. J Invertebr Pathol, 106: 179-191. doi:10.1016/j.jip.2010.09.003. [ Links ]
Delgado, G.A., C.T. Bartels, R.A. Glazer, N.J. Brown-Peterson and K.J. McCarthy. 2004. Translocation as a strategy to rehabilitate the queen conch (Strombus gigas) population in the Florida Keys. Fish. Bull., 102: 278-288. [ Links ]
Espejo, R.T. and J. Romero. 1997. Bacterial community in copper sulfide ores inoculated and leached with solution from a commercial-scale copper leaching plant. Appl. Environ. Microbiol., 63: 1344-1348. [ Links ]
Espejo, R.T ., C.G. Feijoo, J. Romero and M. Vásquez. 1998. PAGE analysis of the heteroduplexes formed between PCR-amplified 16S rRNA genes: estimation of sequence similarity and rDNA complexity. Microbiology, 144: 1611-1617. [ Links ]
García Gaona., M., M.A. Márquez. and C.X. Moreno Herrera. 2016. Characterization of bacterial diversity associated with calcareous deposits and drip- waters, and isolation of calcifying bacteria from two Colombian mines. Microbiol. Res., 182: 21-30. doi:10.1016/j.micres.2015.09.006. [ Links ]
Gerçe, B., T. Schwartz, C. Syldatk and R. Hausmann. 2011. Differences Between Bacterial Communities Associated with the Surface or Tissue of Mediterranean Sponge Species. Microb. Ecol ., 61: 769-782. doi:10.1007/s00248-011-9802-2. [ Links ]
Gram, L. 1993. Inhibitory effect against pathogenic and spoilage bacteria of Pseudomonas strains isolated from spoiled and fresh fish. Appl. Environ. Microbiol ., 59: 2197-2203. [ Links ]
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95-98. [ Links ]
Hammer, Ø., D.A. Harper and P.D. Ryan. 2001. PAST: paleontological statistics software package for education and data analysis. Paleontol. Electr., 4:1-9. [ Links ]
Hammer, Ø ., D.A. Harper and P.D. Ryan . 2005. PAST: Palaeontological statistics, ver. 1.35. Paleontología Electrónica 4. [ Links ]
Hansen, G.H. and J.A. Olafsen. 1999. Bacterial Interactions in Early Life Stages of Marine Cold Water Fish. Microb. Ecol ., 38: 1-26. doi:10.1007/s002489900158. [ Links ]
Hovda, M.B., B.T. Lunestad, R. Fontanillas and J.T. Rosnes. 2007. Molecular characterization of the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquaculture, 272: 581-588, doi: 10.1016/j.aquaculture.2007.08.045. [ Links ]
Jayatilake, G.S., M.P. Thornton, A.C. Leonard, J.E. Grimwade and B.J. Baker. 1996. Metabolites from an Antarctic sponge-associated bacterium, Pseudomonas aeruginosa. J. Nat. Prod., 59: 293-296. [ Links ]
Kindinger, L.M., P.R. Bennett, Y.S. Lee, J.R. Marchesi, A. Smith, S. Cacciatore, E. Holmes, J.K. Nicholson, T.G. Teoh and D.A. MacIntyre. 2017. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome, 5: 6. doi:10.1186/s40168-016-0223-9. [ Links ]
Kirjavainen, P.V. and G.R. Gibson. 1999. Healthy gut microflora and allergy: factors influencing development of the microbiota. An. Med., 31: 288-292. [ Links ]
Kisand, V. and J. Wikner. 2003. Combining Culture-Dependent and -Independent Methodologies for Estimation of Richness of Estuarine Bacterioplankton Consuming Riverine Dissolved Organic Matter. Appl. Environ. Microbiol ., 69: 3607-3616. doi:10.1128/AEM.69.6.3607-3616.2003. [ Links ]
Klussmann-Kolb, A. and G.D. Brodie. 1999. Internal storage and production of symbiotic bacteria in the reproductive system of a tropical marine gastropod. Mar. Biol., 133: 443-447. [ Links ]
Kowalik, G., M. Davis, A. Shawl, R.A. Glazer , G.A. Delgado and C. Evans. 2006. Metamorphic response of queen conch (Strombus gigas) larvae exposed to sediment and water from nearshore and offshore sites in the Florida Keys.: Proc. Gulf Caribb. Fish. Inst., 717-729. [ Links ]
Lalloo, R., S. Ramchuran, D. Ramduth, J. Görgens and N. Gardiner. 2007. Isolation and selection of Bacillus spp. as potential biological agents for enhancement of water quality in culture of ornamental fish: Isolation and selection of Bacillus spp. as potential biological agents. J. Appl. Microbiol., 103: 1471-1479. doi:10.1111/j.1365-2672.2007.03360. [ Links ]
Lalloo, R ., D. Maharajh, J. Görgens and N. Gardiner . 2008. Functionality of a Bacillus cereus biological agent in response to physiological variables encountered in aquaculture. Appl. Microbiol. Biotechnol., 79: 111-118. doi:10.1007/s00253-008-1403-8. [ Links ]
Lan, A., A. Bruneau, C. Philippe, V. Rochet, A. Rouault, C. Hervé, N. Roland, S. Rabot, and G. Jan . 2007. Survival and metabolic activity of selected strains of Propionibacterium freudenreichii in the gastrointestinal tract of human microbiota-associated rats. Br. J. Nutr., 97: 714. doi:10.1017/S0007114507433001. [ Links ]
Landínez-García, R.M., J.D. Rangel-Medrano, E.R. Castro-González y E. Márquez. 2011. Variación genética temporal del caracol pala (Strombus gigas) evidenciada por microsatélites en el atolón Bolívar, Archipiélago de San Andrés, Providencia y Santa Catalina. Cuadernos del Caribe, 14: 75-82. [ Links ]
Li, M., H. Yang and J.D. Gu. 2009. Phylogenetic Diversity and Axial Distribution of Microbes in the Intestinal Tract of the Polychaete Neanthes glandicincta. Microb. Ecol ., 58: 892-902. doi:10.1007/s00248-009-9550-8. [ Links ]
Lin, K.-Y., S. Y. Sheu, P. S. Chang, J. C. Cho and W. M. Chen. 2007. Oceanicola marinus sp. nov., a marine alpha proteobacterium isolated from seawater collected off Taiwan. Int. J. Syst. Evol. Microbiol., 57: 1625-1629. doi:10.1099/ijs.0.65020-0. [ Links ]
Márquez, E., R.M. Landínez-García, S.P. Ospina-Guerrero, J. Aicardo, M.P. Segura, E. Castro, J.L. Correa and C. Borda. 2012. Genetic analysis of queen conch Strombus gigas from South West Caribbean. Proc. Gulf Car. Fish. Inst., 114-121. [ Links ]
McCracken, V.J., J.M. Simpson, R.I. Mackie and H.R. Gaskins. 2001. Molecular ecological analysis of dietary and antibiotic-induced alterations of the mouse intestinal microbiota. J. Nutr., 131: 1862-1870. [ Links ]
McKnite, A.M., M.E. Perez-Munoz, L. Lu, E.G. Williams, S. Brewer, P.A. Andreux, J.W.M. Bastiaansen, X. Wang, S.D. Kachman, J. Auwerx, R.W. Williams, A.K. Benson, D.A. Peterson, and D.C. Ciobanu. 2012. Murine Gut Microbiota Is Defined by Host Genetics and Modulates Variation of Metabolic Traits. PLoS ONE, 7: e39191. doi:10.1371/journal.pone.0039191. [ Links ]
Merrifield, D. and E. Ringø. 2014. Aquaculture nutrition: gut health, probiotics, and prebiotics. Chapter. The gut microbiota of fish, 4:75-94. [ Links ]
Miller, E.A., J.A. Livermore, S.C. Alberts, J. Tung and E.A. Archie. 2017. Ovarian cycling and reproductive state shape the vaginal microbiota in wild baboons. Microbiome, 5: 8. doi:10.1186/s40168-017-0228-z. [ Links ]
Mohammadi, S.A. and B.M. Prasanna. 2003. Analysis of genetic diversity in crop plants-salient statistical tools and considerations. Crop Science, 43: 1235-1248. [ Links ]
Nakagawa, H., M. Sato and D.M. Gatlin. 2007. Dietary supplements for the health and quality of cultured fish. Chapter. Microorganisms, 7: 94 - 100. [ Links ]
Navarrete, P., F. Magne, C. Araneda, P. Fuentes, L. Barros, R. Opazo, R. Espejo and J. Romero . 2012. PCR-TTGE Analysis of 16S rRNA from Rainbow Trout (Oncorhynchus mykiss) Gut Microbiota Reveals Host-Specific Communities of Active Bacteria. PLoS ONE , 7:e31335. doi:10.1371/journal.pone.0031335. [ Links ]
Nei, M. and W.H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A ., 76: 5269-5273. [ Links ]
Ó Cuív, P., D. Aguirre de Cárcer, M. Jones, E.S. Klaassens, D.L. Worthley, V.L.J. Whitehall, S. Kang, C.S. McSweeney, B.A. Leggett and M. Morrison. 2011. The Effects from DNA Extraction Methods on the Evaluation of Microbial Diversity Associated with Human Colonic Tissue. Microb. Ecol ., 61: 353-362. doi:10.1007/s00248-010-9771-x. [ Links ]
Ogier, J.-C., O. Son, A. Gruss, P. Tailliez and A. Delacroix-Buchet. 2002. Identification of the Bacterial Microflora in Dairy Products by Temporal Temperature Gradient Gel Electrophoresis. Appl. Environ. Microbiol ., 68: 3691-3701. doi:10.1128/AEM.68.8.3691-3701.2002. [ Links ]
Pappalardo, R. and N. Boemare. 1982. An intracellular Streptococcus, causative agent of a slowly developing disease in the Mediterranean crab, Carcinus mediterraneus. Aquaculture, 28: 283-292. [ Links ]
Pérez, O.M., M. Posada Elorza, G.E. Cadavid Restrepo and C.X. Moreno Herrera . 2014. Assessment of the bacterial community diversity associated with the queen conch Strombus gigas (Linnaeus, 1758) from the Caribbean coast of Colombia using denaturing gradient gel electrophoresis and culturing. Aquacult. Res., 45: 773-786. doi:10.1111/are.12016. [ Links ]
Prada, M., E. Castro, E . Taylor, V. Puentes, R. Appeldoorn and N. Daves. 2008. Non-detrimental findings for the Queen Conch (Strombus gigas) in Colombia, in: NDF Workshop Case Studies WG. https://cites.org/sites/default/files/ndf_material/WG9-CS3-S.pdf. [ Links ]
Rademaker, J.L. and F.J. De Bruijn. 2008. Section 7 update: Computer-assisted analysis of molecular fingerprint profiles and database construction: 1397- 1446. In: Molecular Microbial Ecology Manual. Springer. 3299-3347. [ Links ]
Rawls, J.F., B.S. Samuel and J.I. Gordon . 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U.S.A ., 101: 4596-4601. [ Links ]
Reed, S.E. 1996. Reproductive anatomy and biology of the genus Strombus in the Caribbean: I. Females. Proc. Gulf Car. Fish. Inst., 44: 413-426. [ Links ]
Reid, G., J.A. Younes, H.C. Van der Mei, G.B. Gloor, R. Knight and H.J. Busscher. 2011. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. rev. Microbiol., 9: 27-38. doi:10.1038/nrmicro2473. [ Links ]
Reveco, F.E., M. Øverland, O.H. Romarheim and L.T. Mydland. 2014. Intestinal bacterial community structure differs between healthy and inflamed intestines in Atlantic salmon (Salmo salar L.). Aquaculture , 15: 262-269. doi:10.1016/j.aquaculture.2013.11.007. [ Links ]
Riquelme, C., G. Hayashida, N. Vergara, A. Vasquez, Morales and P. Chavez. 1995a. Bacteriology of the scallop Argopecten purpuratus (Lamarck, 1819) cultured in Chile. Aquaculture , 138:40-60. [ Links ]
Rodriguez, A.I., H. Hariharan and S. Nimrod. 2011. Occurrence and antimicrobial drug resistance of potential bacterial pathogens from shellfish, including Queen Conchs (Strombus gigas) and Whelks (Cittarium pica) in Grenada. Wedmed Centr. Microbiol., 2: WMC001943. [ Links ]
Romero, J. and P. Navarrete. 2006. 16S rDNA-Based Analysis of Dominant Bacterial Populations Associated with Early Life Stages of Coho Salmon (Oncorhynchus kisutch). Microb. Ecol ., 51: 422-430. doi:10.1007/s00248-006-9037-9. [ Links ]
Rungrassamee, W., A. Klanchui , S. Maibunkaew, S. Chaiyapechara, P. Jiravanichpaisal and N. Karoonuthaisiri . 2014. Characterization of Intestinal Bacteria in Wild and Domesticated Adult Black Tiger Shrimp (Penaeus monodon). PLoS ONE , 9: e91853. doi:10.1371/journal.pone.0091853. [ Links ]
Saitou, N. and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4: 406-425. [ Links ]
Sambrook, J. and D.W. Russell. 2002. Molecular Cloning: A Laboratory Manual 3rd ed. Chapter 6. Isolation of DNA Fragments from Polyacrylamide Gels by the Crush and Soak Method. Cold Spring Harbor Laboratory Press, New York, NY, USA. 76:484-487. [ Links ]
Sanguinetti, C., E. Dias Neto and A. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques, 17:915-919. [ Links ]
Sfanos, K., D. Harmody, P. Dang, A. Ledger, S. Pomponi, P. McCarthy and J. Lopez. 2005. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Syst. Appl. Microbiol., 28: 242-264. doi:10.1016/j.syapm.2004.12.002. [ Links ]
Smith, B., S. Bodé, B.L. Petersen, T.K. Jensen, C. Pipper, J. Kloppenborg, M. Boyé, K.A. Krogfelt, and L. Mølbak. 2011. Community analysis of bacteria colonizing intestinal tissue of neonates with necrotizing enterocolitis. BMC Microbiol., 11: 1. [ Links ]
Sommer, F. and F. Bäckhed. 2013. The gut microbiota - masters of host development and physiology. Nat. Rev. Microbiol., 11: 227-238. doi:10.1038/nrmicro2974. [ Links ]
Spor, A., O. Koren and R. Ley, R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol ., 9: 279-290. doi:10.1038/nrmicro2540. [ Links ]
Stoner, A.W. 1996. Queen conch, Strombus gigas, in fished and unfished locations of the Bahamas: effects of a marine fishery reserve on adults, juveniles, and larval production. Fish. Bull ., 94: 551-564. [ Links ]
Stoner, A.W . and M. Ray-Culp. 2000. Evidence for Allee effects in an over-harvested marine gastropod: density-dependent mating and egg production. Mar. Ecol. Prog. Ser., 202: 297-302. [ Links ]
Stoner, A.W . and J.M. Waite. 1991. Trophic biology of Strombus gigas in nursery habitats: Diets and food sources in seagrass meadows. J. Molluscan Stud., 57: 451-460. [ Links ]
Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G.R. Gibson , M.D. Collins and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol ., 65: 4799-4807. [ Links ]
Sun, Y.Z., H.L. Yang, R.L. Ma, K. Song and W.Y. Lin. 2011. Molecular analysis of autochthonous microbiota along the digestive tract of juvenile grouper Epinephelus coioides following probiotic Bacillus pumilus administration: Probiont modulates gut microbiota. J. Appl. Microbiol ., 110: 1093-1103. doi:10.1111/j.1365-2672.2011.04967.x. [ Links ]
Tamura, K., J. Dudley, M. Nei and S. Kumar. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol ., 24:1596-1599. [ Links ]
Tewfik, A. 1997. Life history, ecology, fisheries, stock status, and management measures of the queen conch, Strombus gigas. CARICOM Fish. Res. Doc., 19:84-117. [ Links ]
Thaiss, C.A., N. Zmora, M. Levy and E. Elinav. 2016. The microbiome and innate immunity. Nature, 535: 65-74. doi:10.1038/nature18847. [ Links ]
Trabal, N., J.M. Mazón-Suástegui, R. Vázquez-Juárez, F. Asencio-Valle, E. Morales-Bojórquez and J. Romero . 2012. Molecular analysis of bacterial microbiota associated with oysters (Crassostrea gigas and Crassostrea corteziensis) in different growth phases at two cultivation sites. Microb. Ecol ., 64: 555-569. doi:10.1007/s00248-012-0039-5. [ Links ]
Wright, E.S., L.S. Yilmaz and D.R. Noguera. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol ., 78: 717-725. doi:10.1128/AEM.06516-11. [ Links ]
Yu, D., G. Wang, J. Xie, S. Guan, Z. Hu and L. Wu. 2007. Activity change of protease and amylase in digestive organs of grouper, Epinephelus coioides. Journal of Zhejiang Ocean University (Natural Science), 26: 246-251. [ Links ]
Yúfera, M. and M.J. Darías. 2007. Changes in the gastrointestinal pH from larvae to adult in Senegal sole (Solea senegalensis). Aquaculture , 267: 94-99. doi:10.1016/j.aquaculture.2007.02.009. [ Links ]
Zheng, L., X. Han, H. Chen, W. Lin and X. Yan. 2005. Marine bacteria associated with marine macroorganisms: the potential antimicrobial resources. An. Micobiol., 55: 119-124. [ Links ]
Received: October 05, 2017; Accepted: July 09, 2018











 text in
text in