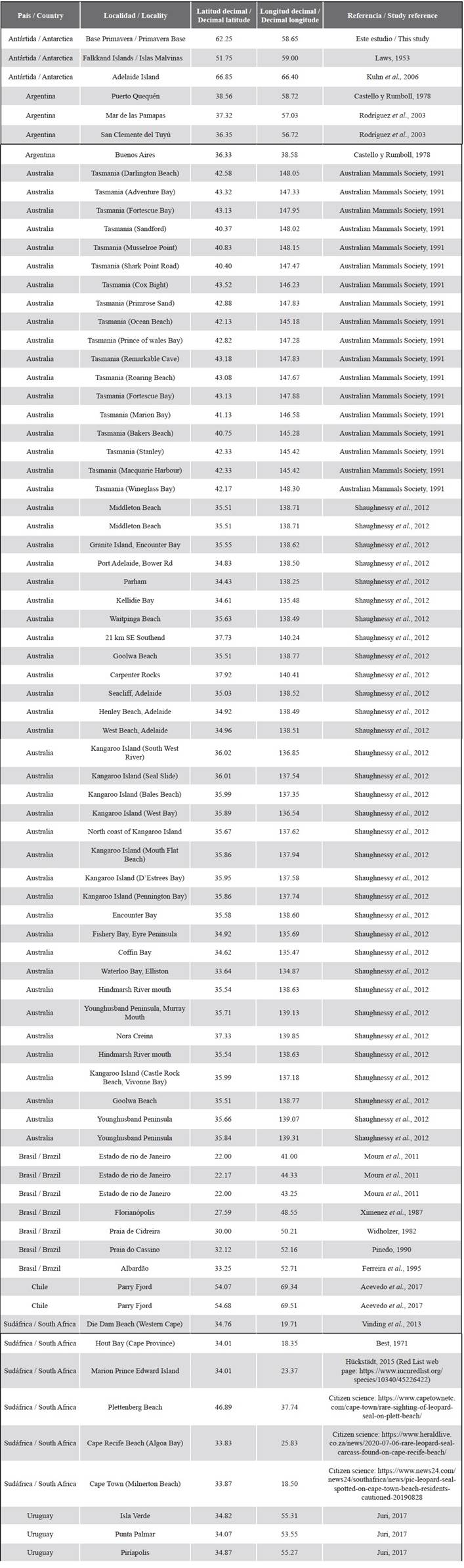
The leopard seal (Hydrurga leptonyx) is an Antarctic pinniped with a wide circumpolar distribution that includes both Antarctic and Subantarctic waters (Bester et al., 2002; Rodríguez et al., 2003; Hückstädt, 2015; Rogers, 2018) (see Figure 1). The leopard seal is a solitary and pagophilic species that disperses widely at low densities along the ice shelf (Rogers, 2009), and some individuals move to Subantarctic zones. Opportunistic sightings of leopard seals in several countries outside their primary range include Argentina (Castello and Rumboll, 1978; Rodríguez et al., 2003; Daneri et al., 2011), Brasil (Pinedo, 1990; Rosas et al., 1992; Ferreira et al., 1995; Silva, 2004; Moura et al., 2011), Chile (Hückstädt, 2015; Acevedo et al., 2017), Uruguay (Juri, 2017), South Africa (Best, 1971; Vinding et al., 2013; Hückstädt, 2015; unpublished data from citizen science), New Zealand, and Australia (Shaughnessy et al., 2012; Hückstädt, 2015; Rogers, 2018) (Figure 1, table S1 in supplementary material). However, this species shows area-restricted behaviors (Meade et al., 2015) and as a result, it may potentially show some population structure (Davis et al., 2008).
Leopard seal shows a wide range in diet and feeds on prey from low trophic level such as krill, to high trophic level such as fish, cephalopods, penguins, and even pups of other seal species (Hall-Aspland and Rogers, 2004; Zhao et al., 2004; Casaux et al., 2009; Botta et al., 2018). Climate change has caused a reduction in ice levels, which has subsequently changed the epipelagic plankton assembly (Mendes et al., 2013; Giraldo et al., 2019; Mojica and Franco-Herrera, 2019). Consequently, leopard seals have shown increased consumption of krill (Guerrero et al., 2014; Botta et al., 2018). These dietary specializations could show lead to long-term population genetic divergences as has been reported in Antarctic killer whales (LeDuc et al., 2008).
Divergences related with life history traits of this species have been reported in vocalizations emitted by leopard seal individuals (Thomas and Golladay, 1995; Rogers and Cato, 2002), which are mainly associated with reproductive behavior (Rogers, 2018). Particularly, adult males tend to have more unique vocal repertoires (Rogers and Cato, 2002; Rogers, 2007), which could have repercussions on the population’s structure if this acoustic divergence affects partner selection by females (Rogers, 2009).
Given the leopard seal’s biology traits as area-restricted behaviors, divergences in diet and vocalizations related to reproduction, and fidelity to molting and reproduction sites (Walker et al., 1998; Hiruki et al., 1999; Hall-Aspland et al., 2004; Negrete et al., 2014), it is possible that there is some degree of genetic differentiation among individuals (Hamilton, 2009). Nevertheless, because the leopard seal’s pagophilic habits allows an expanded distribution onto the pack ice, it is also possible that individuals can keep genetic connectivity between isolated populations. However, genetic analyses of the leopard seal are very scarce (e.g., Slip et al., 1994; Fyler et al., 2005; Bonillas-Monge, 2018) due to its wide solitary distribution even in remote areas (Southwell et al., 2003; Davis et al., 2008), making it one of the least studied species of Antarctica’s phocids at the population level. Therefore, here we present the first description of the genetic diversity of leopard seals in the Danco Coast, near Base Primavera in the Antarctic Peninsula, using a 423 base pairs (bp) fragment of the maternally inherited mitochondrial DNA Control Region (mtDNA-CR).
Blood samples of 13 adult leopard seals were collected during the austral summer of 2011-2012 near the Base Primavera (62º15’S, 58º39’W) at the Danco Coast (64° 09’ S, 60° 57’ W, Figure 1). The animals were immobilized using a dart launch system with a tele-injection air gun from either the ice or from a boat depending on the individuals’ location (Botta et al., 2018). Sex determination was performed by visual inspection of the external genital organs. Blood samples were taken from extradural intervertebral vein and stored in EDTA (ethylenediaminetetraacetic acid) at -20 °C for subsequent genetic analyses.
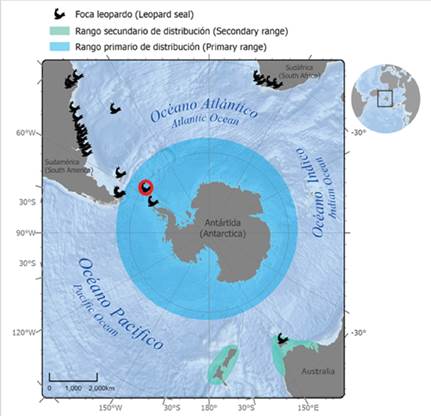
Figure 1 Distribution range of the leopard seal (Hydrurga leptonyx) on the Antarctic Peninsula as primary range, including occurrences outside Antarctic waters as secondary range (Oceania, South Africa, and South America). Red circle indicates location of Argentine Antarctic Base Primavera (62º15’S, 58º39’W), which is located in the vicinity of the Danco Coast northwest of the Antarctic Peninsula (64°09’S, 60°57’W), where leopard seal samples were collected for this study.
DNA was extracted using the Stratec molecular© kit for blood and tissue following standard blood extraction protocol. A fragment of approximately 450 bp of mtDNA-CR was amplified by a Polymerase Chain Reaction (PCR) using the primers forward TRO (5-CCTCCCTAAGACTCAAGG-3’) and reverse D (3’-GTAGACCAAGAATGAAGTCC-5’) (Slade et al., 1994), following the protocol described in Barragán-Barrera et al. (2017). The PCR products were purified following the Polyethylene glycol protocol (20 % PEG), and the DNA was sequenced using Sanger’s method (Sanger and Coulson, 1975).
Sequences were trimmed to 423 bp, edited and aligned using the Muscle algorithm in the software Geneious version 11 (Drummond et al., 2009). Haplotype characterization was performed using MacClade’s software (Maddison and Maddison, 2011). The software Network (Bandelt et al., 1999) was used to build a haplotype network with sequences obtained and the only mtDNA-CR sequence available in NCBI for the leopard seal published by Slade et al. (1994) (GenBank accession number U03590.1), using the Median Joining (M-J) method. Finally, the software Arlequin 3.5 (Excoffier and Lischer, 2010) was used to assess genetic diversity including the haplotypic (h) and nucleotide (π) diversity indexes.
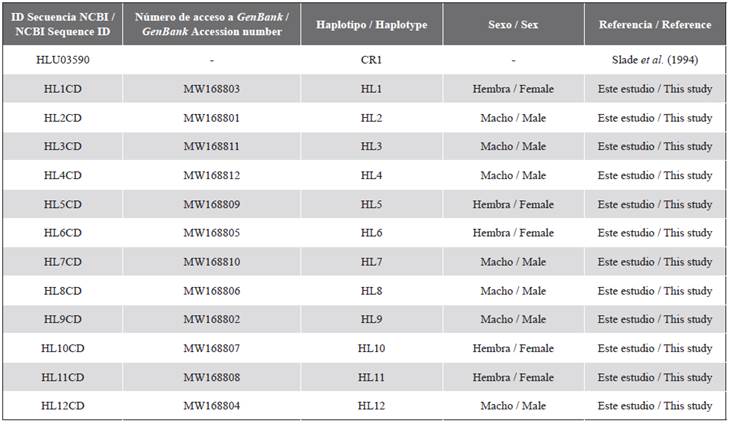
Thirteen haplotypes were found in the 14 analyzed individuals (12 haplotypes from 13 Danco Coast samples, and one haplotype reported by Slade et al. (1994), of which only the haplotype named HL6 was shared by two individuals. The 12 haplotypes sequences identified in Danco Coast were submitted to GenBank as accession numbers MW168801 to MW168812 (table S2 in supplementary material).
In the haplotype network, the haplotype that was reported by Slade et al. (1994), named here as CR1, was connected by one mutational step to haplotype HL1, which appears to be the most ancestral, due to their central position and the number of connections to other haplotypes originating in it (Caballero et al., 2015). Haplotypes HL2, and HL3 were also connected to HL1 by one mutation step, while haplotypes HL7, HL8, HL9, HL10, and HL12 were connected to HL1 by more than one mutational step (Figure 2). For example, HL7, HL8, HL10, and HL12 appear to be the more divergent haplotypes, separated from HL1 by a higher number (> 5) of mutational steps.
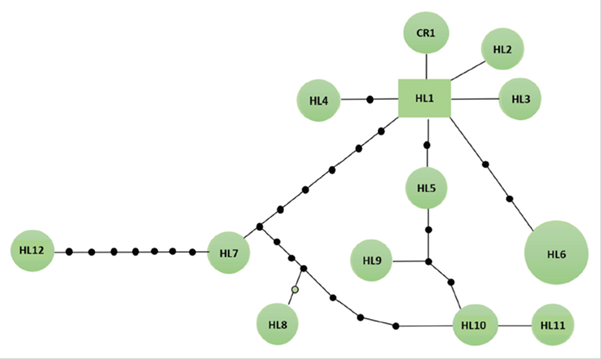
Figure 2 Haplotypic network of leopard seals individuals (Hydrurga leptonyx) presented in the Danco Coast, Antarctic Peninsula. CR1 haplotype corresponds to the unique published sequence in NCBI by Slade et al. (1994) .
Regarding haplotype (h) and nucleotide (π) diversities, in this study we found high values for the leopard seal (h = 0.99; π = 1.76 %; N = 14), similar to values reported for other pinniped species that reproduce on pack ice such as the hooded seal (Cystophora cristata; h = 0.98; π = 1.47 %; N = 123; Davis et al., 2008). These pagophilic species show high genetic diversities likely due to the unstable nature of ice habitat, which hinders site fidelity, resulting in a large reproductive population. In fact, it has been suggested that pagophilic pinniped distribution at low densities along pack ice does not favor the development of population structure even if individuals return to natal areas (Davis et al., 2008).
Our results suggest that the leopard seal population sampled near the Primavera Base has a high genetic diversity, represented by a very low number of shared haplotypes (except for the HL6 haplotype), as well as some divergent haplotypes, which is particularly interesting considering the low sample size and that all the individuals were sampled in the same area. It is possible that leopard seals sampled in the Danco Coast could represent different phylogroups, likely because this species shows site fidelity to reproductive sites despite to its wider distribution favored by their pagophilic habits (Rogers, 2018). However, it is possible that climate change may affect its distribution and potentially its genetic structure, particularly in the western Antarctic Peninsula where higher temperatures have been reported (Vaughan et al., 2003; IPCC, 2014; Mojica-Moncada et al., 2019). Likely, more ice pack presence can maintain a wider genetic connectivity or can restrict gene flow due to premature thawing ice pack. Further genetic studies including the evaluation of nuclear genes and the inclusion of a higher number of samples from other geographical regions along the Antarctic Peninsula is necessary to determine the genetic status of the leopard seal in this region, given its potential vulnerability to the effects of climate change.











 text in
text in