1. Introduction
This paper proposes a strategy for prioritizing electronic medical records (SPIRE) using structured analysis and natural language processing techniques to improve precision in patient selection required for retrospective studies or administrative analysis. This goal is motivated by the increasing adoption of health information systems in primary care, which has produced millions of electronic medical records EMRs all over the world [1]. While medical professionals and institutions are under increased pressure to adopt or further exploit EMR systems, evidence exists of persistent productivity loss [2]. More importantly, no significant evidence exists of improvement on patient outcomes [3]. Nonetheless, at the same time, EMR benefits are clear in terms of decision-making quality, adherence to guidelines, increased revenues or clinical research [2]-[4]. As such, EMR system implementation continues to be complex, and its adoption is still not a clear-cut decision in all cases [5].
The inclusion of unstructured, narrative or textual data further complicates the issue. While implementing a structured and codified EMR system (through forms, ontologies or classifications) may result in expected improvements on information quality, no positive impact has been proven on healthcare professionals or patients [6]. The use of narrative text in EMRs is motivated by the nature of medical care and the need to fully describe unfolding events and facts related to each patient that are not adequately recorded in structured attributes at discharge and include the final diagnosis, secondary diagnoses or co-morbidities, which are rarely coded by many health institutions. Moreover, users must make a personal choice regarding usability, depth or flexibility in narrative accounts, both for entry and retrieval [7].
Text mining and big data technology are significant enablers for obtaining equilibrium among the codification effort, the EMR usability and utility in the presence of already large amounts of narrative data, and the use of data analytics, including data mining. Existing approaches to exploit free text contained in the EMRs for research or quality assessment often rely on manually reviewing a record sample [8]-[10]. Furthermore, EMR analysis for administrative purposes typically relies exclusively on the information contained in structured attributes, neglecting valuable information contained in narrative text [11]. Some studies have proposed strategies for annotating the EMR narrative text using information retrieval and text mining techniques [12]. This kind of strategy is useful for adding value and speeding the search of unstructured text attributes, but by requiring annotation it relies on an additional step or process before actual exploitation even begins. In addition, the studies do not place the annotation process in the context of the EMRs themselves, which could make use of the already structured attributes to guide the tagging or annotation more accurately. Other existing proposals focus on a disease and their extension to other diseases is not straightforward [13]-[15].
As a consequence, a strategy for prioritizing EMRs using structured analysis and natural language processing techniques was designed to improve precision when patients are selected for retrospective clinical studies or administrative analysis, although in this paper we test only for patient identification. This strategy, dubbed SPIRE, enables a collaborative definition of the patient characteristics required for a study; for instance, patients with a diagnosis that have a set of demographic characteristics and have received a treatment with a new medicine. Using the information in both narrative and structured attributes on the EMR system, it prioritizes the records that match such requirements. SPIRE recognizes the EMRs of patients who have been diagnosed with a particular condition, even when this information is not explicitly present in structured attributes, but instead is found in free text, which is often the case (for instance, EMRs may have the initial diagnosis as a structured attribute and the final diagnosis in free text at the end). SPIRE was implemented in a software product that allows its usefulness and precision to be validated with different types of chronic diseases. Given the research group’s expertise, an initial set of three chronic diseases was chosen for validation: diabetes mellitus, pulmonary hypertension and heart failure.
In the following, Section 2 presents the analysis of related works on free text and medical record analysis. Section 3 presents SPIRE in detail, including the proposed method of structuring the requirements around the records necessary for a study. Section 4 details SPIRE’s results applied to different types of studies and diseases. Finally, Section 5 concludes this paper and proposes future work.
2. Background and Related Work
Natural language processing (NLP) and text mining have been used to exploit and analyze EMR free text either for specific diseases or for more generic uses. NLP stems from probabilistic modeling, information theory, and linear algebra [16], as well as from machine learning models to improve narrative text understanding [17]. Text mining is the process of extracting interesting information and knowledge from unstructured text [18]. Text mining uses NLP tasks during the pre-processing phase, which gives structure to the originally unstructured text. Classical NLP tasks include part-of-speech tagging, named entity recognition, disambiguation, and evaluation of semantic similarity.
NLP applications using EMRs, or text mining, have been shown to improve disease knowledge. One study uses NLP and classification models to identify reportable cancer cases from clinical notes [10]; it generates a list of cases that can be validated more easily by final users. Similar proposals analyze characteristics and patterns of patients with alopecia areata [19], asthma [13], diabetes [14], cancer [15], uveitis [20], or cataracts [21], among others. These proposals are focused on one disease at a time and use the text contained in progress notes as input. Our approach is generic, meaning medical experts can create disease- or population-related conditions at any time.
Other applications demonstrate NLP’s utility for identifying clinical events irrespective of the disease, such as postoperative complications [22], falls [14], or adverse drug reactions [23], [24]. These systems work for specific, pre-configured patterns of behavior or against a set of existing quantitative indicators. Again, our proposal allows for the configuration of search conditions that may change over time without being limited to an information need or event.
In addition to medical information needs, other proposals aim to analyze administrative data and reports to assess complaints [25], emergency room use [26] or quality measures [27]. While these systems also apply NLP techniques, they do not match this analysis to medical knowledge bases or corpora because they are centered on administrative data. Our approach, while focused on clinical data inside the EMRs, is also useful for identifying patients for administrative analytics.
An additional set of related works provide complementary or supporting services for EMR preparation. This set includes a large set of works related to anonymization or de-identification to remove confidential or sensitive information from the text and preserve privacy [28]-[31]. Even though the goal of anonymization proposals is not to analyze or prioritize EMRs, the NLP techniques applied to recognize identifiers and quasi-identifiers are very close to the process of identifying entities for other purposes. Other existing research for EMR text pre-processing has demonstrated the possibility of extracting temporal expressions [32], [33]; correcting misspelled words [34]; resolving existing co-references [35]; eliminating redundancy [36] and generating summaries [37].
Finally, software tools, such as Electronic Medical Record Search Engine (EMERSE) [38] and Queriable Patient Inference Dossier (QPID) [39], have provided automated methods for acquiring semantics from free text structures in EMRs. A wide array of EMR, clinical business intelligence and analytic tools are commercially available and have begun to incorporate more sophisticated and intelligent query and analytical services. The main difference to our approach is that, while others require the institution to replace their systems, our strategy can be applied over any EMR system.
All the above related works focus on analyzing narrative text without combining the analysis with other (structured) fields within the record. They assume that the records to be analyzed have already been identified (i.e., they already have a list of patients with a condition), or they require the hospital or institution to change its current EMR system. Our proposal aims to advance the analysis of medical records, providing a general system that combines the analysis of narrative texts and structured attributes, allowing the collaborative definition of search conditions, without requiring the hospital or institution to change its current system or the healthcare professionals to change their EMR input practices.
3. Method: SPIRE-A Strategy for Prioritizing Electronic Health Records
This section presents SPIRE, a strategy for searching and prioritizing EMRs, using structured analysis and natural language processing techniques to improve precision in selecting patients required for retrospective clinical studies or administrative analysis. SPIRE involves a collaborative definition of the patient characteristics required for a study (Section 3.2), and, using the information embedded in narrative and structured attributes in the EMR system (Section 3.3), it prioritizes the records that best match such characteristics (Section 3.4). The premise behind SPIRE is that even if some of the information contained in EMRs is well structured (such as medication dosage or laboratory test orders), some events are hidden in narrative text -like comorbidities, demographic characteristics or risk factors- which are also valuable for different kinds of analysis.
This section presents an overview of SPIRE, followed by an account of how search templates are defined and then used to analyze structured and unstructured data with prioritized results.
3.1. SPIRE Overview
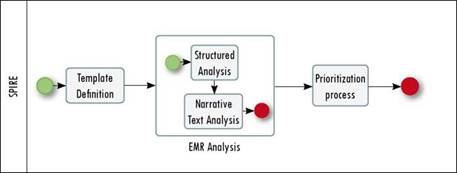
The goal of SPIRE is to increase the number and relevance of patients identified for a study, maintaining a positive predictive value. To do that, SPIRE combines socially driven collaboration, natural language processing techniques, structured analysis and a prioritization algorithm that allow the required EMRs to be obtained. Figure. 1 presents the general process followed by SPIRE.
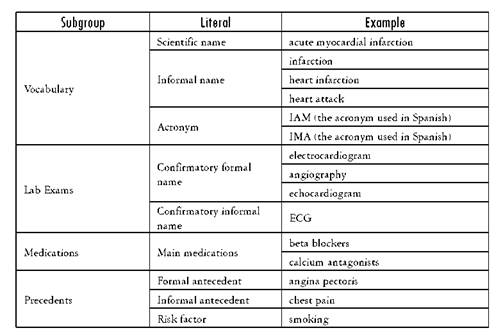
The starting point of SPIRE is the definition of a search template, according to the information needs of a researcher or analyst (or team), as shown in the Template Definition (Figure 1) and described in Section 3.2. The template is divided into n subgroups S that are composed of m literals L (see Definition 1). An example of a subgroup is Vocabulary and its literals are Scientific name, Formal name, Informal name, Synonyms and Acronyms.Table 1 shows a template example; the first column represents the subgroups used to describe the required records, the second one includes the literals used to specify each subgroup, and the last column is an example of the possible values of the literals. Values need not be included for all the literals or all the subgroups in a template, only for those required for the analysis.
Definition 1. Template. A Template D is composed of a set of subgroups S={s 1 ,s 2 , ..., s n } that describe the main characteristics of the required EMRs. Each subgroup s i is specialized by patient and is composed of a set of literals L(s i ) = {l i1 , l i2 , ..., l im }, where l ij represents a fixed value for an atomic characteristic of the EMRs.
The information contained in the template is then used to parameterize the analysis over the EMRs (see EMR Analysis in Figure 1). The analysis is divided into two phases, an initial phase uses structured information, and the second uses NLP to explore narrative text (e.g., medical notes, progress notes, and nursing notes). Section 3.3 presents both phases, resulting in a set of matching records.
Finally, SPIRE uses the analytical output to prioritize the records that better match the template requirements (see Prioritization Process in Figure 1). The prioritization method is detailed in Section 3.4.
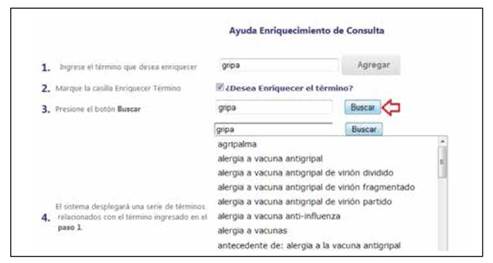
The Vocabulary and Medications subgroups can be enriched using the terms provided by SPIRE related to an initial term defined by the user. Figure 2 presents an example of the concept “gripa” (flu in English) and a set of the related terms in Spanish that can be used to improve the template. The underlying terminology used for this enrichment uses a clinical vocabulary employed in a national hospital. For the medications, SPIRE also enriches automatically the term using the available commercial terms or the generic formula.
The first group of tests using SPIRE allowed us to conclude that lab exams used to confirm a diagnosis (confirmatory exam) are better for improving the strategy’s specificity and sensitivity than the word describing the diagnosis per se. For this reason, we include this group as a literal.
3.2. Collaborative EMR Search Templates
SPIRE centers on patient identification for research and/or administrative purposes, so each query differs. Sometimes the purpose is to collect all patient records related to a disease or with a diagnostic test or treatment. Other times, the query may be generated to obtain all EMRs related to a service or department within the hospital. As such, SPIRE is not meant to work as a traditional search engine. Rather, given the highly specialized and codified nature of EMR data, it combines structured and non-structured attributes in the query that improve the results’ relevance. In addition, to generate and test search templates, it supports an iterative and collaborative approach. Through this process, the template structure can be refined and a stable set of characteristics for which to look can be obtained, either individually or by sharing the template (and subsequent versions) with other users. Each template contains the following groups: vocabulary (associated terms), precedents (personal and family history), lab exams, medications, symptoms, treatment plans and exclusion terms (terms which may negate the results, such as “discard” or “not found”).
However, this process goes beyond mere template creation. When working on our original three cases for chronic diseases, it became clear that by iterating different versions of the template not only were we arriving at a satisfactory description with which to structure the analysis and its prioritization but also useful knowledge was being generated in the templates themselves. The templates are stored in the systems and contribute to a collaborative knowledge creation process among users.
As a result, the choice was made to incorporate this collaborative template building process into the system. Each time a researcher or administrative staff member wishes to query a set of conditions in the EMR system, they have two template options: private or public. A private template is an individual query that can be saved for exclusive use. This process provides each user not only with a history of past queries executed in the system but also with the ability to refine the query as the user sees fit. Thus, each query template is named, identified and potentially versioned so it can evolve towards an ever more precise description of the information need, which can then be reused and/or altered in the future.
When a public template is created, the effect becomes collaborative. For instance, the initial contribution may be named in relation to a medical condition. By being public, the user invites others to evaluate and use this template. The first effect is that the effort that goes into designing a template -given that determining exactly what to look for in each case is often an expert-dependent knowledge-intensive contribution- can be reused by other staff interested in the same type of query. Once the template is incorporated into this shared knowledge base, other users may simply run it again (perhaps changing the date range or patient gender).
The second benefit of public templates is that they can be socially evaluated, enriched or revised. If another user finds an improvement (by adding, changing or removing attributes in the template), they can create a new version of the template without affecting the previous one. This functionality of collaboratively creating and refining EMR search templates creates the net effect of supporting collective intelligence and knowledge management, giving SPIRE the ability not only to provide faster and more accurate search results but also to create a repository in which medical and administrative personnel have a shared space to represent, store and share codified, highly specialized and contextual knowledge.
3.3. Structured and Non-Structured Analysis
Once the query template is created, SPIRE executes the analysis to obtain the EMRs required by a user. By combining structured variables and text, the program increases the narrative text’s context using the structured information in the EMRs. For this analysis, SPIRE divides each EMR M into a set of p structured attributes S and a set of q narrative text attributes T (see Definition 2).
Definition 2. Electronic Medical Record. An electronic medical record M is composed of a set of structured attributes C = {c 1 , c 2 , ...,c p } whose value domain is discrete and a set of narrative text attributes T = {t 1 ,t 2 ,...,t q } whose domain is a natural language text.
The goal of the analysis is to detect within C k and T k of an EMRs’ M k , the value of each of the literals lij pertaining to the analyzed template (see Definition 1).
The principle is to find the majority of literals described in the template within each EMR, regardless of whether they are contained in a structured or a narrative text attribute. When the values found in the EMRs are more literal, they will have greater emphasis in the final result.
Currently, SPIRE includes two search functions that detect the occurrence of literal values as follows: one detects a literal’s value in structured attributes C and another searches within narrative text attributes T. Additional search functions can be added according to other attributes. For example, a search function could be added that would analyze attributes related to images.
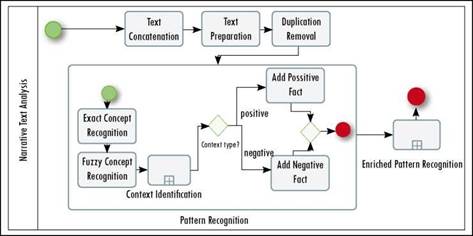
Searching structured attributes is straightforward using classic structured query language (SQL) queries. In contrast, searching within narrative texts includes a preparation of texts and analysis using natural language processing methods. These methods include the recognition of entities and the disambiguation of entities that are written negatively. For example, although the phrase “discard diabetes” includes an important entity, it is used in a negative sense. Figure 3 illustrates the phases that SPIRE executes as it analyzes narrative texts.
Because each medical record has different free-text fields, the first task is to concatenate all the texts for one patient (i.e., Text Concatenation) to maintain a correct historical precedence of facts. The complete text is then prepared using classical NLP tasks such as sentence splitting, tokenization and POS (Part of Speech) recognition (i.e., Text Preparation). Through sentence splitting, SPIRE identifies sentences within the text; then, tokenization divides the text into small units using space and punctuation marks as dividing characters; the POS process identifies each token’s grammatical function within the sentence. These tasks are performed using GATE architecture [40], [41]. In some situations, the exact text of a note is used in another note (i.e., copy/pasteproblem), so the analysis continues by removing all the duplicate paragraphs or sentences that may exist in the complete text (i.e., Duplication Removal). This task may be optional if the EMR system already controls this problem. Once the text is integrated and cleaned, SPIRE recognizes patterns according to the concepts defined in the Template. This recognition includes searching the literal in the text (i.e., Exact Concept Recognition), even if the text contains some misspellings (i.e., Fuzzy Concept Recognition). The context of the recognized literal is analyzed using near negative and positive concepts (i.e., Context Identification). If the number of negative concepts found exceeds a threshold, the pattern is categorized as a negative one. This task uses a knowledge base of terms and expressions previously classified as positive or negative. This knowledge base contains terms such as “discard”, and expressions such as “there is no evidence” classified as negative. In [42] we propose an approach for named entity recognition and concept attribute labeling for EMRs that considers the contextual words around the entity of interest to determine its sense. The final task performs the same actions described previously, but it changes the concepts of the templates with to synonyms included in the knowledge base (i.e., Enriched Pattern Recognition).
The following algorithm describes the steps executed during the analysis. This algorithm uses the EMR set as the input and returns them prioritized according to the query criteria. This process includes the query execution over the EMR system in a traditional sense, using structured attributes and textual analysis extracted from the same system. This algorithm works in a general sense over the EMR set and does not require training or learning for conditions. Steps (1) and (2) are executed simultaneously.
Input: Template D, EMRSet {EMR 1 , EMR 2 , ..., EMR n }
Output: EMRPrioritizedSet {(EMR 1 , p 1 ), (EMR 2 , p 2 ), ..., (EMR n , p n )}
Begin
Matrix M = {}{} //The matrix that will contain the results
Matrix Ms = {}{} // The matrix that contains the structured analysis result
Matrix Mt={}{} // The matrix that contains the text analysis result i = 0, j = 0
ForAll (s in D) // for each group in the template query= BuildSQLQuery (L(si)) // Build a group of SQL queries using the literals defined in the subgroup i
ForAll (EMR in EMRSet)
(1) sResult = ExecuteQuery(query,EMRj) If (sResult !=null) // If at least one literal of the template subgruoup is found in the EMR using structured analysis Update (Ms{i}{j},sResult)
End_if
(2) PText = PreprocessingText(EMRi) // Concatenate, prepare and eliminate redundancies tResults = TextAnalysis(PText, L(si)) //Semantic Named Entity Recognition If (t Result!=null) // If at least one literal of the template subgroup is found in the EMR using text analysis Update (Mt{i}{j},tResult)
End_if
Update (M {i} {j}, integration (tResults, sResults)) j++ End ForAll i++
End ForAll
Return prioritizedEMR(M{i}{j})
End
SQL queries included in the algorithm extract information from EMR structured attributes. The most common of these are diagnoses, prescribed and supplied medications, medical labs and procedures. The diagnosis attribute uses the ICD10 classification [43]. SQL queries are built dynamically according to the literals defined in the template.
At the end of the search process, the output is the result for each literal, including the number of hits for each one of them and a negative mark for the literals found in a negative context. These results are used as inputs in the prioritization algorithm described in the following section.
3.4. Prioritization
Consider a template asking for patients who have pulmonary emphysema, who are receiving a treatment with inhaled steroids, who had a tomography scan and who have obesity. After applying the analysis, SPIRE may return 10 patients with all these characteristics, 6 with all but the obesity, and 5 with everything but the treatment. One alternative is to provide only the first 10; however, some of the other patients may be useful for the study, and the decision to skip or maintain them requires human analysis. For this reason, we include a prioritization algorithm that integrates the results following semantic rules related to the template definition. For instance, when replaceable attributes in the template are found in different parts of the record, as in the case of the disease’s formal and informal names, the integration process increases this record’s importance with respect to the attribute. Following semantic rules, integration is used to calculate the distance between the template’s description (original query) and the record (the obtained EMR). The EMR with the shorter distance corresponds to a higher match against the template (e.g., the 10 patients who match everything defined in the template) (see Section 3.5).
The integration process considers Definitions 3-5.
Definition 3. Subgroup Intensity. The intensity I of a subgroup of literals s. is the normalized percentage of matched literals within the EMRs’ M j . If a literal has multiple possible values (e.g., multiple acronyms) each value is considered a literal (e.g., Acronym 1, Acronym 2, etc.).
Definition 4. Subgroup Utility. The utility U of a subgroup of literals s. is a percentage value of its importance in identifying an EMR’s template, assuming that all the literals’ values are positive.
Definition 5. Subgroup Level of Hits. The number of hits H of a subgroup s. is the normalized number of times that literal values were matched within M j .
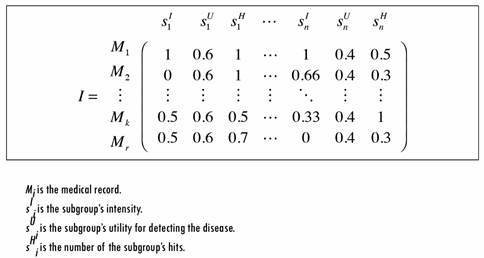
The intensity reflects the percentage of matched literals within an EMR for each subgroup. This value is important for prioritizing records with respect to a subgroup. To calculate each subgroup’s utility, a classic method of multi-criteria decision analysis is used in which each subgroup is evaluated on multiple criteria by experts, and the utility is “the average specified in terms of normalized weightings for each criterion, as well as normalized scores for all options relative to each of the criteria” [44]. The number of hits is used as an optional calibration value that considers the number of times that literal values are found in an EMR. The intention is to assign a higher weight to records in which the same literal repeats. This value is optional because for some subgroups the number of repetitions is important (e.g., Vocabulary), but for others it is not (e.g., Family Diseases). Figure 4 illustrates an example of the integration process output.
As seen in Figure 4, each of the rows represents an EMR and each set of three columns represents the analysis results for one subgroup of the query template. For example, for the first EMR’s M1 the matrix concludes as follows: All the literals defined in the first subgroup S1 were found (i.e., s I 1 = 1), either in the text or in a structured attribute.
The found literals have the highest number of repetitions compared with the other EMRs analyzed. The equation s u 1 = 1 represents the normalized number of hits in the EMRs.
The subgroup has a utility of 0.6 when SPIRE evaluates the prioritization. This value is the same for all the records in this subgroup.
3.5. Distance Function
The distance function between the template T and each analyzed EMR’s M can be calculated using any distance function, such as Euclidean or Cosine distance. The template description and each EMR are represented in an n-space, where n is the number of subgroups.
The record with the shortest distance is the first one in the prioritized list; records with a distance of one are discarded, and others are organized according to the distance value. EMRs that were marked as having negative literals are marked with a negative sign before the priority value.
3.6. Validation
SPIRE was validated by comparing the results of its automated searches with manual searches performed by a general practitioner in a random sample of 396 EMRs corresponding with patients hospitalized in internal medicine wards. These records are stored in the EMR system of a general hospital with an installed capacity of 350 beds.
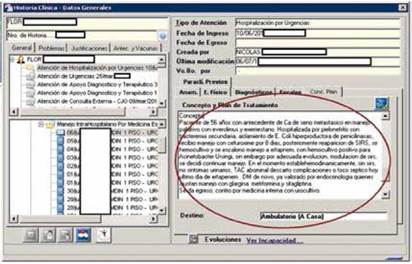
The general practitioner first read each of the 396 EMRs, checking each of them, including narrative text and structured fields, with supervision from an expert in the three chronic diseases selected: diabetes mellitus, pulmonary hypertension and heart failure. After an EMR was manually read, it was categorized as having or not having the disease. The final decision was considered the gold standard for evaluating the behavior of SPIRE in prioritizing the EMRs. Figure 5 illustrates an example of an EMR read by a practitioner. The red circle shows one of the narrative texts contained in the EMR. Although the EMRs include a structured attribute called diagnosis, most of the time, it contains only the admission diagnosis, which may differ from the discharge diagnosis found in the narrative text. This problem was the main motivator for this research.
A software tool that implements the SPIRE strategy, called DISEarch [45], enabled the general practitioner to define query templates for each disease.
3.6.1. Statistical analysis
In the SPIRE automated searches, we identified the following factors included in each template: vocabulary (associated terms), vocabulary taxonomy, precedents (personal and family history), diagnostic test results, medications, symptoms, treatment plans and exclusion terms. We compared these factors with the results obtained from the manual evaluation using cross tabulations to assess the accuracy of each characteristic in detecting a valid diagnosis. We calculated the areas under the ROC curve, with their respective 95% intervals of confidence, and the corresponding sensitivity and specificity values for each of the three diseases: heart failure, pulmonary hypertension and diabetes mellitus. We used logistical regression to identify the best multivariable function in each disease. Considering the goal of creating a single model able to identify different pathologies, we created a latent variable from a number of common covariates, finding the best weights for each with the goal of maximizing the area under the ROC curves. Data were analyzed using Stata, version 11 [45].
4. Results
Of the initial 396 EMRs included in the validation sample, the manual search found 92 patients positive for diabetes mellitus, 62 with pulmonary hypertension and 101 heart failure cases. The validity of specific variables for a correct diagnosis is presented in Tables 2-4. These tables present the results for each of the subgroups defined in the template and for each of the diseases selected for validation in the area under the curve. The variables associated more on univariate analysis with the final outcomes were Vocabulary, confirmatory testing and precedents.
Table 2 Area under the ROC curve with its corresponding 95% confidence interval as well as the sensitivity and specificity for heart failure
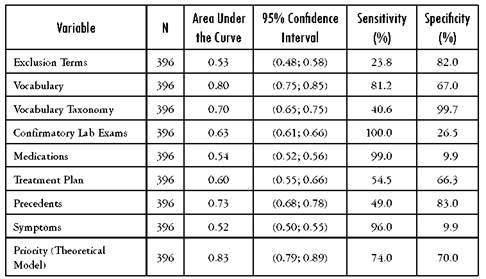
Source: Author's own elaboration
Table 3 Area under the ROC curve with its corresponding 95% confidence interval as well as the sensitivity and specificity for primary pulmonary hypertension
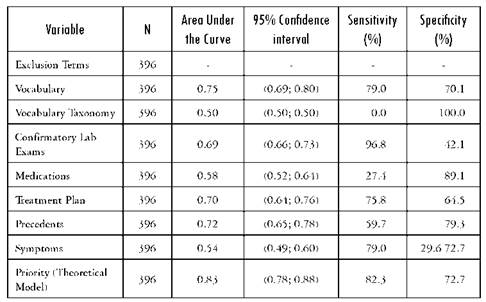
Source: Author's own elaboration
Table 4 Area under the ROC curve with its corresponding 95% confidence interva as well as the sensitivity and specificity for diabetes mellitus
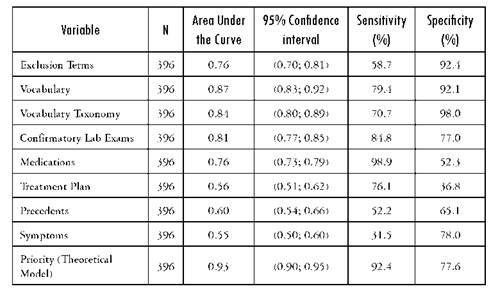
Source: Author's own elaboration
Once we identified the principal variables from the original models, we included all these in a single multivariable model and evaluated its contribution individually. The variables Vocabulary and Precedents were highly correlated, so only Vocabulary was retained in the model. A summary of the variables included in the final diagnostic algorithm and its contribution to the model is presented in Table 5.
Table 5 Variables found important and their final contributions to the multivariable model
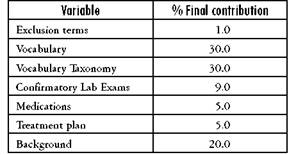
Source: Author's own elaboration
Diagnostic Algorithm: ExclusionTerms 0.01 + Vocabulary 0.30 + Taxono- myVocabulary 0.30 + ConfirmatoryLabExams 0.09 + Drugs 0.05 + Treatment 0.05 + Precedents 0.20
These results allow us to conclude that when a user is defining a new template, the subgroups that have greater importance for recognizing the required EMRs are the Vocabulary and its taxonomy, as well as the Precedents.
This model was used in each of the three diseases, estimating the area under ROC curves with their respective 95% confidence intervals for sensitivity and specificity. The results are presented in the Table 6.
Table 6 Area under the receiver operating characteristic curves (AUC): Comparison of manual search diagnosis and SPIRE software for detecting heart failure
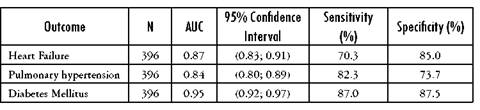
Source: Author's own elaboration
The strategy demonstrated high sensitivity when it was evaluated in each of our three disease models (heart failure, pulmonary hypertension and diabetes mellitus) (70%, 82% and 87%, respectively) and even higher specificity (85%, 73.7% and 87.5%, respectively). The AUC was between 0.84 and 0.9, showing great discrimination ability.
In addition to the previous validation, we compared the results obtained with SPIRE against the results obtained executing queries with structured attributes (e.g., diagnosis) and full-text queries with text-based attributes (e.g., nursing notes). The aim of this test was to compare the results’ precision against SPIRE’s with more than 50% priority.
Specifically, we randomly selected 18 699 EMRs filtered for adults that had been hospitalized within the same time frame. At the same time, we defined SPIRE templates that included only literals on the vocabulary and its specialization. In this test we did not analyze sensitivity or specificity because that process would require reading each EMR against the gold standard. In addition, we expected many true negatives, but the emphasis was on detecting true positives. Nevertheless, we manually validated all the positive results obtained through SPIRE and queries.
The results were conclusive in the sense that SPIRE demonstrated better precision, between 88.1% and 95%; whereas the queries obtained a precision of between 54.4% and 67.5%.
5. Conclusions and future work
This paper presented SPIRE, a strategy that enables researchers, clinicians and hospital administrators to easily search and identify relevant medical records of patients with a diagnosis and other important characteristics for research and decision-making. It was designed to search both structured and unstructured (narrative) data and to produce a prioritized list of EMRs that meet a given criteria. The user sets the criteria using a collaborative template builder, and SPIRE uses natural language processing and a ranking algorithm to prioritize the search results.
This functionality of collaboratively creating and refining EMR search templates creates the net effect of supporting collective intelligence and knowledge management, allowing SPIRE to provide faster and more accurate search results. It also creates a repository in which medical and administrative personnel have a shared space to represent, store and share codified and contextual knowledge. To date, we have tested the sensitivity, specificity and precision of our approach and are monitoring its practical use in a hospital setting to report on usability, collaboration effects and generalizability beyond our first three cases. SPIRE provides a priority value for each EMR, but this provision serves as an indication of compliance to be confirmed or rejected by medical experts; it does not replace them.
The need to create a template for each new search is an advantage because it enables reuse. For example, the template helps to identify infrequent cases to be included in descriptive studies (case series, retrospective cohorts, or case controls studies) or potential candidates for participation in clinical trials. Additionally, the template could be useful for identifying patients for administrative analysis regarding costs or for audit purposes (e.g., adherence to clinical practice guidelines). The main disadvantage is that the tool’s performance depends on the complexity of the query. We dealt with this problem using a variety of diseases in the learning phase to generate our algorithm. For validation purposes we selected three chronic diseases (diabetes mellitus, pulmonary hypertension and heart failure), searching simultaneously in the same population of EMRs, in which the patients had been hospitalized in internal medicine wards. Our results demonstrated that even if sensitivity and specificity were slightly different for each disease, discrimination ability was great in all cases, with AUC values between 0.84 and 0.9. This result was not affected by the presence of comorbidities in the same records.
SPIRE is generally useful for EMR search and prioritization but was designed with an emphasis on chronic diseases, given that each EMR will comprise several entries and events. At this time, SPIRE was not compared against tools or algorithms designed for conditions or contexts. Moreover, while SPIRE was tested for acute or ambulatory conditions or procedures, the results were validated systematically for other diseases or administrative queries. These are interesting questions to be addressed in future work.