Introduction
The inchi (Caryodendron orinocense K.) species belongs to the Euphorbiaceae family, which is comprised of around 60 genera and 529 species, being one of the largest and most diverse families in flowering plants (USDA, 2014). It is known in different parts of the world as metohuayo (Peru); inchi, cacay, tacay (Colombia); cacay, nambi, maní de árbol (Ecuador); palo de nuez, Barinas nut (Venezuela) and castanha do porco (Brazil). It is a species originating in the western Amazon and it is widely distributed in the Amazon Basin in Colombia, Ecuador, Peru and Venezuela (Martinez, 1996). The in-chi tree grows to 15 to 20 meters high and it is a rustic species with great adaptability to acidic soils that have a high aluminum content. It is also considered to be one of the most promising species of the Colombian Amazon and Orinoco Region (Jaramillo and Jaramillo, 2010).
The average production of the adult tree is 250 kg/ year, which represents a family income of COP 200,000/year (Diaz and Ávila, 2002). As well as the use of its trunk for wood, its stone fruit for fuel and its nut for food, the inches most important use is for oil production from the seed. This makes it an excellent oil plant, competing highly advantageously with others such as the African palm, which is used for consumption (Omega 3, 6 and 9), as well as for the manufacturing of cosmetics and pharmaceutical products (Barrio, 2005; Jiménez and Bernal, 2002). The inchi's great potential is not only due to the quality of its oil, but also because of the byproduct from the extraction of the seed, which can be used in animal feed, because it has a high percentage of protein (43-46%) and minerals such as calcium and phosphorous (Tamayo, 1963).
There are limited studies on the morpho-agronomic characterization of inchi. Only the studies of the Instituto Sinchi conducted by Ávila and Cárdenas (2000) have been reported, who assessed the germoplasm of five Amazon species. There are also some still unpublished, preliminary studies by Corpoica (Colombian Corporation for Agricultural Research). Taking into account that knowledge of the genetic diversity is required for the conservation and genetic improvement of the species, there are no genetic or molecular characterization studies, not even on similar species such as Caryodendron amazonicum Ducke, Caryodendron angustifolium Stanley and Caryodendron grandifolium. Within the Euphorbiaceae family, we find the pine nut (Jatropha curcas L.) and sacha inchi (Plukenetia volubilis L.) as species that also produce high-quality oil, on which research has been conducted, designed to genetically identify the materials (Basha et al. 2007; Ganesh et al. 2008; Basha et al. 2009; Jubera et al. 2009; Pamidimarri et al. 2009 a and b; Subramanyam et al. 2009; Cai et al. 2010; Ikbal et al. 2010; Vargas 2011; Corazon et al. 2009; Rodriguez et al. 2010).
Molecular markers known as random amplified microsatellites (RAMs), also known as ISSR (inter-simple sequence repeat) are useful for measuring the genetic diversity of plants and animals, as well as differences between families and species and within species (Muñoz et al. 2008). They show the same basis of variation of individuals, and they allow specific regions in the DNA molecule to be selected for specific studies. The number of detectable polymorphisms is theoretically unlimited and they allow analysis of information that is expressed, as well as unexpressed information (Mahuku et al. 2002; Morillo, 2005). This methodology is feasible for small laboratories in terms of equipment and cost facilities and because it does not require prior knowledge of sequences or require the use of radioactive isotopes. The markers obtained by RAMs may be used for population studies (Hantula et al. 1997).
The objective of this research work was to characterize the genetic diversity of inchi (Caryodendron orinocense K.) using random amplified microsatellites (RAMs) to be able to propose strategies in the future for conservation and genetic improvement of the species.
Materials and Methods
Plant Material
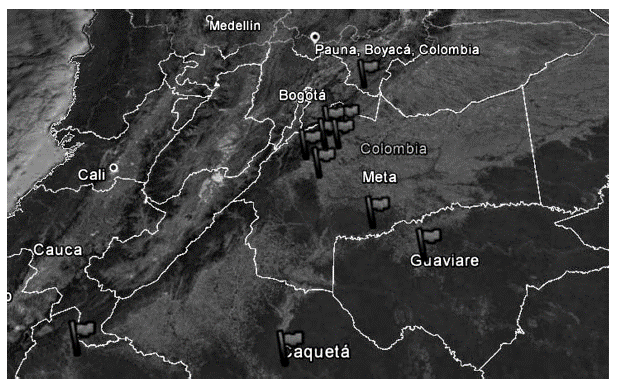
Young leaves were collected from 29 male and female inchi (Caryodendron orinocense K.) trees from the Meta Department in the municipalities of San Juan de Arama, Lejanías, Cubarral, Guamal and El Castillo, Restrepo, and some materials were provided by the KA-HAI S.A.S. nursery (Table 1). A greater amount of plants with female or hermaphrodite sexual organs was included, which ensures high fruit production, as well as good agronomic and adaptability characteristics.
Table 1 Accessions of Caryodendron orinocense used for molecular characterization with RAMS.
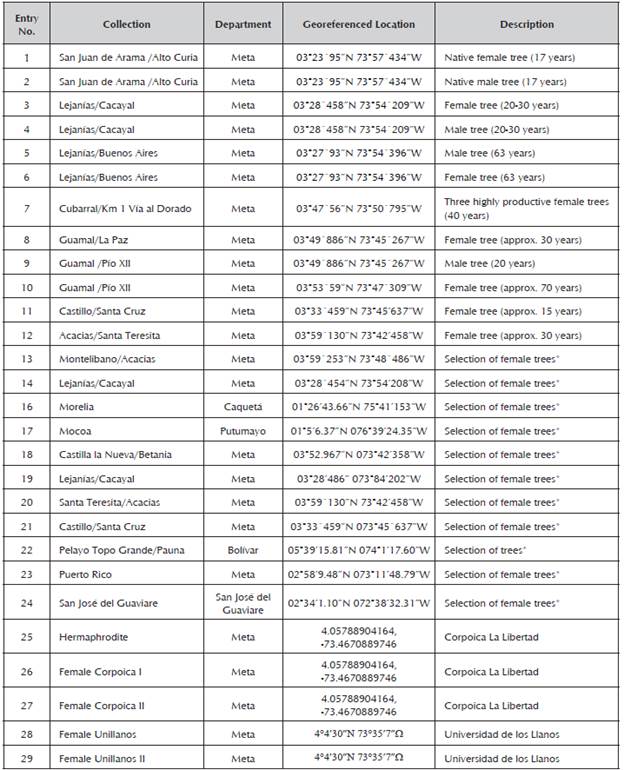
*KAHAI S.A.S.
In Figure 1, the spatial distribution of the Caryodendron orinocense trees used in this study can be observed.
Molecular Characterization
The molecular characterization of inchi was conducted in the plant biotechnology laboratories of the Universidad de los Llanos, located in Villavicencio, Meta, Colombia; with an altitude of 467 m.a.s.l. and an average temperature of 26 °C. The Dellaporta Protocol (1983) was used for the extraction of DNA.
The genomic DNA of the total samples was visualized in 0.8% agarose gel, dyed with ethidium bromide in a Maxicell Primo EC-340 Electrophoresis Gel System chamber. To establish the DNA concentration of each genotype, a dilution curve was made with DNA of the Lambda bacteriophage with an initial concentration of 20 ng/ μl and taken to final concentrations of 20, 40, 60, 80 and 100 ng/μl. The quantified DNA was diluted in HPLC water at a total volume of 100 μl at 10 ng/ μl and it was stored at -20 °C.
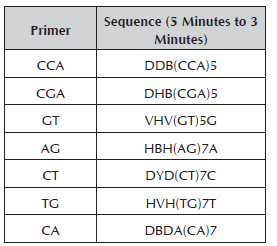
Seven primers synthesized by Technologies Inc. were used for the RAM analysis (Table 2). For the reaction of amplification with RAMs, the cocktail was prepared in a sterile microcentrifuge tube (1.5 ml) for a final volume of 25 μl. The reaction mix was prepared with buffer 1X, MgCl2 1.5 mM, DNTPs 0.2 mM, Taq Polymerase 1U, primer 2 μM and genomic DNA 10 ng.
The amplification was carried out in a PTC 100 Programmable Termal Controller (MJ. Research, Inc). The initial denaturation was at 95 °C for 5 minutes; denaturation at 95 °C for 30 seconds, hybridization at a temperature of 50 °C (primers AG and CA), 55 °C (primers CCA-TG-CT) and 58 °C (primers GT-CGA) for 45 seconds, a final extension of 72 °C for 2 minutes, 37 cycles from denaturation to extension and finally, an extension at 72 °C for 7 minutes. The products were separated by electrophoresis in agarose gels at 1.2% run at 90 volts for 3 hours, visualizing it in an ultraviolet light transilluminator.
Statistical Analysis
A binary matrix of species absence (zero) / presence (one) was generated. The genetic similarity between the individuals was calculated using Nei and Li's similarity coefficient (1979). The cluster analysis was conducted by the UPGMA method and a dendrogram was generated using the NTSYS system statistical package (Numerical Taxonomy System for Personal Computer, Version 2.02 PC). To assess the genetic diversity, the unbiased heterozygosity and the percentage of polymorphic loci were assessed using the TFPGA (Tools For Population Genetic Analyses, Version 1.3, 1997) statistical package. The unbiased f statistic was established with a 95% confidence interval.
Results and Discussion
Out of the 29 inchi genotypes collected, two were rejected because the DNA could not be obtained. The seven RAM primers used for the molecular characterization of 27 genotypes generated a total of 85 bands which contributed 90% to the breakdown of the materials. The number of bands by initiator varied from 10 for the CA primer to 20 for the CGA primer with molecular weights that oscillated between 260 and 1500 Kb (Table 3).
Table 3 Primers used for the molecular characterization of Caryodendron orinocense K, total number of bands and polymorphic bands.
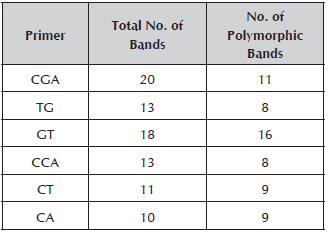
When the number of polymorphic loci (62) found in this study is compared with other work on genetic diversity in species of the Euphorbiaceae family, such as the studies conducted by Vargas (2011) (73 polymorphic bands), Basha and Sujatha (2007) (116 polymorphic bands) and Souza et al. (2009) (27 polymorphic bands) on Jatropha curcas L, the number of bands found is adequate for the estimation of the genetic parameters in inchi. The GT primer was the one that made the greatest contribution to the genetic variation observed, which means that it may be useful to assess the genetic diversity in Caryodendron materials.
The analysis though Nei and Li's coefficient at a similarity level of 0.50 divided the population into 4 groups (A, B, C and D) (Figure 2).
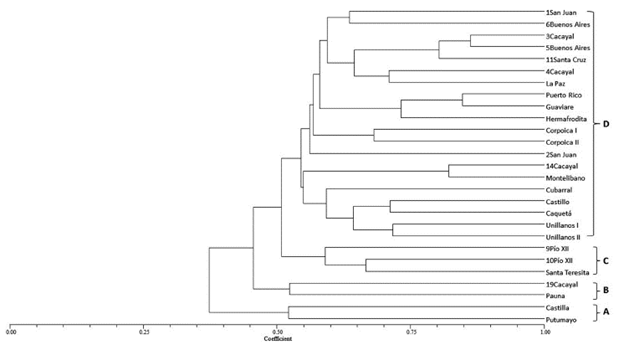
Figure 2 Dendrogram of the genetic structure of 27 individuals of Caryodendron orinocence K based on Nei and Li's similarity coefficient and calculated from the combined data of the seven RAM primers with the UPGMA classification method using the SAHN and TREE programs of NTSYS-pc, Version 1.8.
Groups A and B, which include the materials from Putumayo, Castilla, Pauna and Cacayal 19, are the ones that present the lowest similarity value (0.50) compared to the other groups formed. This can be seen in these materials' own characteristics, such as fruit size, fruit color, leaf size, and color and size of the nut according to observations made by the company KAHAI S.A.S.
As can be observed, Group D brings together the greatest number of materials collected in the Meta Department together with the materials from Guaviare and Caquetá, and it presents a similarity index of more than 0.70. This provides evidence of low genetic variability between the individuals, which may be due to the gene flow, transport of plant material and uniparental origin (FAO, 2002). The materials from Pío XII identified with the numbers 9 and 10 and from Santa Teresita, which comprised Group C, present similar characteristics in terms of their genetic and morphological composition. This could be explained by their geographical proximity, because Guamal and Acacias are municipalities less than 20 km apart.
Generally speaking, the molecular analysis grouped the materials of Caryodendron orinocense according to their geographical distribution, showing that the materials from Putumayo and Boyacá (Pauna) present less genetic similarity than the rest of the evaluated materials, so the materials from Guaviare and Caquetá were grouped with those from the Meta Department. This may be explained by the transport of plant material and the cross-pollination (gene flow) between plants from the same geographical origin.
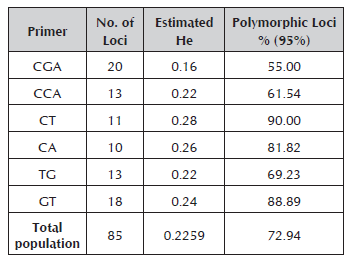
The estimated heterozygosity values were within a range from 0.16 to 0.28 for the CGA and CT primers, respectively. The percentage of polymorphic loci varied between 55% for the CGA primer and 90% for the CT primer (Table 4).
Table 4 Estimated heterozygosity and percentage of polymorphic loci of each one of the primers used for the characterization of genetic diversity in Caryodendron orinocense.
Studies conducted by Vargas (2011) on the identification of genetic diversity in Jatropha using ten ISSR markers found an average polymorphism percentage of 34%, which is similar to that observed in other work on Jatropha, such as the studies by Basha and Sujetha (2007), who evaluated 42 accessions with RAPD and ISSR markers, finding 42% polymorphism and moderate genetic diversity; and Reddy et al. (2007) who using RAPD and AFLP markers in 23 materials obtained 15% and 9% polymorphism, respectively, concluding that the genetic diversity is low, coinciding with the research conducted by Sun et al. (2008). However, other studies show the existence of moderate to high genetic diversity in Jatropha with polymorphism percentages that go from 35% to 98% (Ganesh et al. 2008; Gupta et al. 2008; Pamidiamarri et al. 2009a; Senthil Kumar et al. 2009; Basha et al. 2009; Singh et al. 2010; Tatikonda et al. 2009; and Cai et al. 2010). Studies conducted by Corazon-Guivin (2009) on the genetic characterization of accessions from San Martín from the national germoplasm bank of sacha inchi (Plukenetia volubilis L) show 60.87% of polymorphic loci, which is why the results of this research are found within the results obtained in the studies conducted on the Euphorbiaceae family.
The average fixation index (FST) for the 27 materials studied was 0.35 with a standard deviation of 0.07. According to Wright (1978), values over 0.25 show genetic differentiation. The FST found in this study is an important parameter because it facilitates understanding of the spatial-time dynamics of the materials of Caryodendron, as well as the structure of crosses between them. For the purposes of comparison, Nason (2002) proposes that in agreement with the high rate of exogamy and the dispersion of pollen to large distances that seem to characterize the populations of neotropical trees, said populations present a relatively high diversity and a relatively low degree of genetic differentiation, even in populations that are kilometers apart.
However, in the species studied, despite being alloga-mous, there is a high degree of pollination between related individuals, which results in low genetic diversity and a high fixation index. In the Plukenetia (Euphorbiaceae) genus, Rodríguez et al. (2010) conducted morphological and molecular differentiation studies using ISSR markers, finding a FST between 0.89 and 0.98, with which a high degree of differentiation was shown between the studied accessions. However, it is important to take into account that to study the genetics of populations and their degree of structuring, it is essential to have structured populations and ones with a sufficient number of individuals, as well as measuring the time, allowing conclusions to be drawn on the gene flow and population dynamics.
The greater degree of polymorphism and genetic information contributed by the RAM technique may be complemented with information from morphological and biochemical characterization and therefore, more clearly elucidate the intricate relations and interactions that are presented in the majority of materials and assess their intra-specific diversity on a much finer scale.
Conclusions
RAM markers allow the existing genetic variation in the assessed materials of Caryodendron orinocense to be established, grouping them according to the geographical site where they were collected.
It was determined that there is little genetic diversity, due to uniparental reproduction. However, these materials present genetic differentiation because of the processes of domestication and evolution to which the materials are constantly subjected.











 text in
text in