Introduction
The Rosaceae family is comprised of more than 3,000 species, many of which are fruit trees of economic importance, such as the apricot tree (P. armeniaca L.), almond tree (P. dulcis Mill), the cherry tree (P. avium L.), European plum tree (P. domestica L.) and Japanese plum tree (P. salicina L.) (Fan et al. 2013). The annual production of the most important Prunus species was 76,378,738 (ton) for apples, 23,580,845 (ton) for pears, 21,083,151 (ton) for peaches and nectarines, 3,956,639 (ton) for apricots, 2,256,519 (ton) for cherries and 10,702,744 (ton) for plums (FAOSTAT, 2014). In the Prunus genus, as well as peach (Prunus persicae (L.) Batsch), there are four other different peach species: P. davidiana (Carr.) Franch., P. mira Koehne, P. kansuensis Rehd. and P. ferganensis, but only the P. persica species is grown for its fruit (Hancock et al. 2008). Like the plum (Prunus domestica), they constitute the most popular deciduous species that are cultivated in the temperate zones around the world, as well as the peach. They are a diverse group of plants with many species adapted to an ample range of ecogeographic conditions and that have been grown for centuries (Okie and Hancock, 2008).
The identification of cultivars and the study of genetic relations in this species was traditionally based on morphological characteristics. However, in the Prunus breeding programs, its assessment is a tedious process, due the very long youth period, the influence of the phenological status on the expression of characteristics and the existence of climatic factors that affect its manifestation. Therefore, assisted selection by molecular markers is very useful in these cases (Ercisli et al. 2011). In the Prunus species, the (SSR) microsatellites have been the most used markers for the characterization of germoplasm (Guarino et al. 2009), determination of genetic diversity (Rogatis et al. 2013), handling of germoplasm (Kamm et al. 2011), identification of cultivars and fingerprinting (Gulen et al. 2010), determination of genes of interest (Bouvier et al. 2012), self-incompatibility and population studies (Mariette et al. 2010) and gene mapping (Dhanapal et al. 2012). In Prunus, the microsatellites developed in one species have been used in different species, demonstrating their transferibility and ability to detect polymorphisms (Fan et al. 2013).
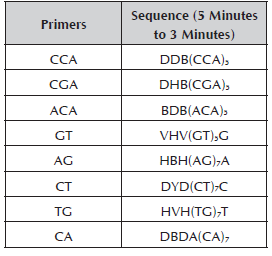
Zietkiewick et al. (1994) proposed a method to measure genetic diversity in plants and animals using primers based on microsatellites. This technique combines the benefits of the microsatellites and RAPD (Random Amplified Polymorphic DNA). Hantula et al. (1996) selected four primers (GT, ACA, CCA and CGA) with a length of 18 bases, which include a 5' "degenerated" end of three nucleotides that serve as an anchor to ensure the union of the primer at the start of the microsatellite. They called this technique RAMs (Random Amplified Microsatellites). RAMs are based on the polymerase chain reaction (PCR). The method is highly reproducible and permits the detection of intra and interspecific polymorphisms in the DNA (Muñoz et al. 2008).
With the RAM markers, populations are studied, genetic diversity in plants and animals is measured, the base of variation of individuals is shown, parts of the genome can be studied, and the information that is expressed is analyzed, because the number of detectable polymorphisms is theoretically unlimited (Mahuku et al. 2002). The methodology can be adjusted for small laboratories in terms of equipment and cost facilities, and it does not require prior knowledge of sequences (Muñoz et al. 2008).
Work on the characterization of genetic plant diversity suggests that the RAM technique is useful for identifying duplicates in germoplasm banks to establish groups or associations between individuals of a population according to the species, geographic location and phylogenetic relations, among others (Muñoz et al. 2008). It is widely used in fruit plants such as Rubus glaucus, where it differentiated the R. glaucus, R. robustus and R. urticifolius species, detected duplicates and found a high genetic variability in R. glaucus, the most important species (Morillo et al. 2005). In the case of Physalis, these markers found high diversity and two accessions of red fruit were genetically differentiated from the yellow fruits (Bonilla et al. 2008). Regarding guava, these primers were highly polymorphic and a high variability was found in Valle del Cauca (Sanabria et al. 2006). In the mandarin, this technique enabled the grouping of accessions according to the satsuma, clementine and hybrid types of mandarin. Therefore, it is a useful tool to measure diversity in the Citrus genus (Morillo et al. 2009; Mora et al. 2013).
Using eight Random Amplified Microsatellites (RAMs), Morillo et al. (2014) measured the genetic diversity of 31 materials of Prunus from the collection of deciduous species of the Universidad Pedagógica y Tecnológica de Colombia. With a similarity coefficient of 0.75, three groups were formed mainly according to the characteristics of the fruit with a heterozygosity value of 0.22. The RAM technique was useful for measuring the genetic diversity in deciduous species.
Within this context, the main objective of this research work was to analyze the interspecific variability of the peach and plum materials with RAM markers of the deciduous species collection of the Universidad Pedagógica y Tecnológica de Colombia to approach the proposal of strategies for hybridization and identification of elite materials.
Materials and Methods
A total of 28 accessions of peach tree, 11 of plum tree, 2 of apricot tree (Prunus armeniaca) and one of almond tree (Prunus dulcis) were evaluated belonging to the collection of deciduous species of the Universidad Pedagógica y Tecnológica de Tunja, planted in the experimental farm of Tunguavita, Paipa, located at 36 km from the city of Tunja with North Latitude of 5° 47'04" and West Longitude of 73°07'17" and an average temperature of 15 °C (Table 1).
Table 1 Prunus material of the deciduous collection of the Universidad Pedagógica de Colombia, used for the evaluation of genetic diversity.

Molecular Characterization
The molecular characterization was conducted in the Molecular Biology, BIOPLASMA and GEBIMOL Research Laboratories of the Universidad Pedagógica de Colombia, Tunja. The Dellaporta et al. (1983) Protocol was used for the extraction of DNA. The total DNA was visualized in 0.8% agarose gel in a Maxicell Primo EC-340 Electrophoresis Gel System chamber. To establish the DNA concentration, the Hoefer Dyna Quant 200 fluorometer was used, it was diluted in HPLC water at a total volume of 100 μl to 10 ng/μl and it was stored at -20 °C. For the RAM analysis, eight primers synthesized by Technologies Inc. Bioneer were used (Table 2). For the reaction of amplification with RAMs, the cocktail was prepared in a sterile microcentrifuge tube (1.5 ml) for a final volume of 25 μl. The reaction mix was prepared with buffer 1X, MgCl2 1.5 mM, DNTPs 0.2 mM, Taq Polymerase 1U, primer 2 μM and genomic DNA 10 ng.
The following designations were used for the degenerated sites: H (A or T or C); B (G or T or C); V (G or A or C) and D (G or A or T).
The amplification was carried out in a PTC 100 Programmable Thermal Controller (MJ. Research, Inc). The initial denaturation was at 95 °C for 5 minutes; denaturation at 95 °C for 30 seconds, hybridization at a temperature of 50 °C (primers AG, ACA and CA), 55 °C (primer CCA-TG-CT) and 58 °C (primer GT-CGa) for 45 seconds, an extension of 72 °C for 2 minutes, 37 cycles of denaturation to extension and finally, an extension at 72 °C for 7 minutes. The amplified products were separated by polyacrylamide gel electrophoresis 37:1 (acrylamide: bisacrylamide) of 7% at 150 v per hour in a small chamber of DNA Sequencing System, FB-SEQ-3545 of Fisher Biotechnologies. It was stained using silver salts following the protocol of Cres et al. (2001).
Statistical Analysis
A binary matrix of species absence (zero) / presence (one) was generated. For the selection of polymorphic bands, a polymorphic locus was considered to be one in which the most common allele frequency was less than 95%. The genetic similarity between the individuals was calculated using Nei and Li's similarity coefficient (1979). The grouping analysis was conducted with the SAHN program of NTSYS-pc (Version 2.02 g, 98) using UPGMA (Unweighted Pair Group Method with Arithmetic Mean). The dendrogram was constructed with the TREE program of NTSYS-pc (Version 2.02 g, 98). To assess the genetic diversity, the unbiased heterozygosity and the percentage of polymorphic loci were assessed using the TFPGA (Tools For Population Genetic Analyses, Version 1.3, 1997) statistical package. The AMOVA® statistical package was used through the AMOVA PREP program to conduct the analysis of molecular variance (AMOVA) and therefore, be able to establish the difference between and within the groups formed. The unbiased f statistic was established with a 95% confidence interval.
Results and Discussion
The analysis through Nei and Li's coefficient at a similarity level of 0.75 differentiated the materials into three groups (Figure 1).
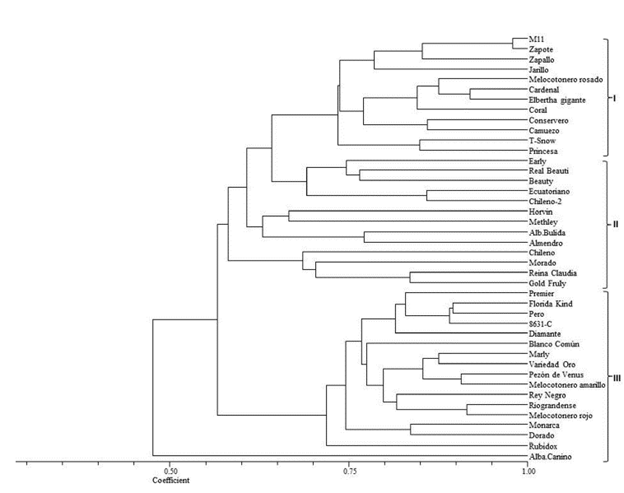
Figure 1 Dendrogram of 42 materials of Prunus based on Nei and Li's similarity coefficient and calculated with eight RAM markers with the UPGMA, SAHN and TREE classification method of NTSYs-pc, Version 1.8 (Exeter Software, NY, USA).
Peaches are classified by the presence of trichomes on the skin and the adherence of the flesh to the pit. Classical peaches (Prunus pérsica L.) have velvety skin and the flesh is attached to the pit. Peaches (Prunus pérsica l.) with velvety skin but with flesh that readily separates from the pit are called freestones. Nectarine/peaches (Prunus pérsica var. nectarina) that have smooth skin and flesh that readily separates from the pit are called "freestone nectarines" (Campos et al. 2013). Taking into account that this classification in Group I contains peach varieties strictly called Conservadero of yellow flesh with red coloring around the pit and a medium size, which was introduced from Ecuador, and the Camuezo variety with yellow flesh and prolific production, it is used as a rootstock for all varieties of peach and some of plum. The Jarillo is a variety imported from Venezuela with yellow skin and flesh and it is used as a rootstock in many nurseries (Campos et al. 2013).
There are more than 15,000 varieties of peach in the world (Castro et al. 1998). In Colombia, the Fresa Morado, Pezón de Venus, Fresa Blanco, Elberta Gigante, Rubidoux, Frank, Ventura, Diamante, Riograndense, Dorado, Conservero Amarillo and Zapallo varieties stand out (Ordoñez, 2007), some of which are found in this group.
A large part of the plums evaluated in this study are found in Group II. According to the color of the fruit's skin, they have been classified in yellow fruits like the Ecuadorian variety, which has a low content of epicuticular wax and propagates vegetatively to obtain root-stocks and graft plum varieties such as Kelsey, Sangre Toro, Duarte and Beauty, which are not compatible with the common peach, the commonly used root-stock. Varieties are classified in red fruits such as the Beauty varieties with epicuticular wax and heart shape, which is the commercially most abundant on the market, although it is very susceptible to post-harvest manipulation; Methley with red flesh and a sweet flavor, which is very prolific, has early production and despite its size, does not have problems on the market; and Horvin, which has red skin with epicuticular wax, is prolific and has early harvest, with it being possible to obtain two harvests a year, mainly in systems with a bimodal rain system (Campos et al. 2013).
The most common rootstock used in plum grafting is the white common peach. However, it presents incompatibilities with some varieties of plum. The white flower plum tree is also used but with the same problems as above. Therefore, they are replaced with Ecuadorian, Red Japanese or myrobalan plums. The latter is characterized for being rustic with a small, red-color fruit and small leaves without any commercial value with fresh fruit. The almond tree is a species that presents good compatibility in grafting with different varieties of plum tree, which is why it is part of this group (Campos et al. 2013).
In Group III, one of the most important peach varieties is the Florida variety, which has adapted to the conditions of the Cauca Region and it is planted at 2,400 m.a.s.l in the municipalities of Silvia and Sotará. Its fruits are of good quality and very juicy with a red color and yellow flesh. Production starts at three years after planting and it can produce fruits for 20 years (Serna and Oliva, 1999). The Dorado and Diamante varieties are characterized by having yellow skin with light red pigmentation and golden-yellow flesh with red coloration around the pit. The former presents good characteristics for industrialization and because of its good sugar content, it is excellent for fresh consumption. The latter is characterized by being a very strong and productive plant. The Oro variety with an attractive yellow color presents good organoleptic properties (Baiza, 2004).
Rubidoux is included in the peach varieties for being resistant to post-harvest handling, having a yellow flesh with red coloring and a lightly round shape, as well as the Rey Negro variety, which has yellow skin with opaque red stains and the presence of a very pronounced tip, causing problems in post-harvest handling (Campos, 2013). The canino apricot was the material that presented the least similarity with the rest of the evaluated material, although greater affinity has been reported with other species of the Prunus genus (Gallego, 2010).
In general terms, the grouping of the peach material evaluated with the RAM markers corresponded to the species evaluated and the fruit characteristics like in other studies of genetic diversity using microsatellite markers in Prunus (Stanys et al. 2008). However, a high degree of similarity is observed between species such as peach and plum, and the latter with apricot, which may be useful for the establishment of hybridization strategies within a genetic breeding program of Prunus.
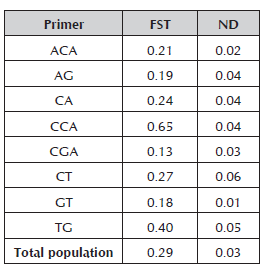
The eight RAM primers used for molecular characterization in peach generated a total of 121 alleles, which fluctuated between nine for the GT primer and 26 for the ACA primer with molecular weights between 260 and 1000 kb. The percentage of polymorphic loci varied between 71% and 98.7% for the CGA and CT primers, respectively (Table 3). The number of alleles is considered to be adequate to estimate genetic parameters when comparing it with the results obtained in other species of Prunus (Stanys et al. 2012; Fernández et al. 2009). The CCA primer was the one that made the greatest contribution to the genetic variation observed, FST 0.65 (Table 4), which means that it may be useful to achieve greater differentiation between the materials of the Prunus genus.
Table 3 Estimated heterozygosity (He), number and percentage of polymorphic loci for the eight RAM primers evaluated with 42 materials of Prunus.
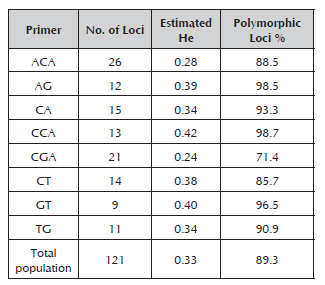
Table 4 Population differentiation, FST statistic for the 42 materials of Prunus evaluated with the eight RAM primers.
The heterozygosity values were between 0.24 for the CGA primer and 0.42 for the CCA primer. The average heterozygosity value for all the evaluated materials was generally 0.33, much lower than those found in genetic diversity studies using SSRs, such as those conducted by Carrasco et al. (2012) on 29 elite cultivars of Japanese plum (Prunus salicina) and four cultivars of Prunus (0.80); Barac et al. (2014) on 35 accessions of Prunus cerasus (0.39); and Font et al. (2013) on 94 cultivars of peaches and nectarines (0.45). However, it was higher than the values obtained by Barac et al. (2014) when evaluating seven accessions of P. mahaleb (0.28), 22 of P. avium (0.31), four of P. fruticose (0.31), four of P. serrulata (0.28) and four of P. persica (0.19).
The low levels of heterozygosity found are attributed to the mating systems, because the majority of the peach cultivars is self-compatible, which favors the self-pollination processes (Sassa et al. 1992). Low levels of polymorphisms have also been found in sweet cherry materials (Boskovic and Tobutt, 1997). This probably reflects a narrow genetic base in the sweet cherry and peach germoplasm. However, it coincides with the statements of Imbert and Lefevre (2003), who consider that an excess of heterozygosity is not very common in tree species and that this is generally promoted by self-incompatibility and interspecific crossings (Stoeckel et al. 2006). The narrow genetic base used for the development of cultivars and the existence of common ancestors in the pedigree peaches and plums could explain the results obtained (Okie and Hancock, 2008). In general, the low levels of variability are consistent with those obtained using different molecular markers in species of the Prunus genus (Aranzana et al. 2010).
The fixation index (FST) obtained when evaluating the 42 materials of Prunus with the eight RAM markers was 0.29 with a standard deviation of 0.03 (Table 4). According to Wright (1978), values of 0.25 demonstrate great genetic differentiation, which may be reflected in the high degree of domestication that these materials have suffered, because the majority of them is comprised of commercial varieties. This value is higher than the one found by Carrasco et al. (2012) in the plum tree (-0.13) and much higher than in other cross-pollination species such as the cherry tree (0.092; Stoeckel et al. 2006), almond tree (0.15; Fathi et al. 2008) and apricot tree (0.04; Bourguiba et al. 2010). The above demonstrates that the peach and plum varieties are less genetically diverse, unlike the results obtained by Font et al. (2013). This may be explained by taking into account the evolutionary history of the different species in their domestication process and clone propagation system (McKey et al. 2010), but also by the fact that the peach is the least polymorphic species of the Prunus species because of its autogamy (Font et al. 2014).
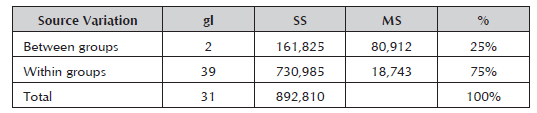
The RAM markers broke down the materials into three groups at very reduced genetic differences (0.75), thus showing the degree of consanguinity that exists between them. Carrasco et al. (2012) found similarity values from 0.57 to 0.99 when characterizing 29 Japanese plum (Prunus salicina) cultivars and four peach cultivars with SSR and ISSR markers. High similarity values have also been obtained between and within the different species of the Prunus genus (Yoon et al. 2006; Fernández et al. 2009). The analysis of molecular variance (AMOVA) for the three groups showed a variance between the groups of 75% (Table 5), and the remaining 25% was due to the genetic variance component within the groups.
The similarity existing between the different materials evaluated supports the idea that there is a high capacity for hybridization and the gene flow between the Prunus species evaluated. The narrow genetic base used for the development of varieties could explain these results, as the materials studied may have a common ancestor within their pedigree (Okie and Hancock, 2008).
The high degree of synteny that arises between the peach and plum genomes, as could be observed in this study, may be used for the production of hybrids, because there are studies that show how these two species are used for the generation of new and better materials within the breeding strategies of the Prunus genera (Frecon, 2006; Reales et al. 2010). The use of molecular markers and the analysis of sequences (Scalabrin et al. 2013; Vicedomini et al. 2013) will allow a more detailed study of the genome of the Prunus species and in turn, it will be a useful tool to add to the strategies of hybridization between the species of the Prunus genus and make them more efficient.
Conclusion
Molecular characterization with RAM markers of the collection of peach and plum trees of the Universidad Pedagógica y Tecnológica de Colombia showed that there is a high degree of similarity between the evaluated materials and therefore, the diversity was low. However, this shows the high degree of consanguinity of the species evaluated and that it can be used efficiently within hybridization systems that lead to obtaining new and better Prunus plants.











 texto em
texto em